Comments (6)
Hi Nelson,
Thanks for trying out our new package. We suspect the issue you are experiencing is due to the passing of a fasta file as a genome object. i.e. this line
genome <- "/media/ot/Data/Nelson/refdata-cellranger-GRCh38-3.0.0/fasta"
Instead you should set the genome object to a BSgenome S4 data object from the appropriate BSgenome package. i.e.
genome <- BSgenome.Hsapiens.UCSC.hg38::BSgenome.Hsapiens.UCSC.hg38
If you don't already have this package you can download it via the following commands:
if (!requireNamespace("BiocManager", quietly = TRUE))
install.packages("BiocManager")
BiocManager::install("BSgenome.Hsapiens.UCSC.hg38")
Let us know if this fixes the problem.
Regards,
Dave
from sierra.
Hi Dave,
Thanks for your prompt reply. It solved the problem.
But when I move on to SplitBam function, No data found. I have tested several genes. Bam file is from the output of CellRanger. What is the problem with my data set?
Regards,
Nelson
SplitBam function:
> str(cells.df)
'data.frame': 1872 obs. of 2 variables:
$ celltype: Factor w/ 4 levels "cartilaginous",..: 1 3 3 3 3 3 1 3 3 2 ...
$ cellbc : chr "AAACCTGGTTAAGGGC" "AAACCTGTCGAGAGCA" "AAACGGGAGAACAATC" "AAACGGGAGAATCTCC" ...
>
> SplitBam(bam = "./L07/possorted_genome_bam.bam",
+ cellbc.df = cells.df,
+ outdir = outdir,
+ gtf_gr = gtf_gr,
+ geneSymbol = "ENG")
splitting bam file: ./L07/possorted_genome_bam.bam
[1] "cartilaginous" "stress" "angiogenic" "fibroblast"
processing cell type cartilaginous
[W::hts_idx_load2] The index file is older than the data file: ./L07/possorted_genome_bam.bam.bai
chunk0: 8192 length of aln: 0
No data found for cartilaginous
processing cell type stress
[W::hts_idx_load2] The index file is older than the data file: ./L07/possorted_genome_bam.bam.bai
chunk0: 8192 length of aln: 0
No data found for stress
processing cell type angiogenic
[W::hts_idx_load2] The index file is older than the data file: ./L07/possorted_genome_bam.bam.bai
chunk0: 8192 length of aln: 0
No data found for angiogenic
processing cell type fibroblast
[W::hts_idx_load2] The index file is older than the data file: ./L07/possorted_genome_bam.bam.bai
chunk0: 8192 length of aln: 0
No data found for fibroblast
Additional scripts:
### Visualization
library(Seurat)
library(stringr)
library(dplyr)
#Previous definitions
out.dir <- "example_TIP_aggregate"
#Read in the counts
peak.counts <- ReadPeakCounts(data.dir = out.dir)
# Correct cell id
colnames(peak.counts) <- colnames(peak.counts) %>% str_split("-") %>% sapply("[[", 1)
#Read in peak annotations
peak.annotations <- read.table("TIP_merged_peak_annotations.txt",
header = TRUE,
sep = "\t",
row.names = 1,
stringsAsFactors = FALSE)
head(peak.annotations)
#Load precompiled gene-level object called 'genes.seurat'
Low_L07 <- readRDS("../scRNAseq/Low_L07.rds")
peaks.seurat <- PeakSeuratFromTransfer(peak.data = peak.counts,
genes.seurat = Low_L07,
annot.info = peak.annotations,
min.cells = 0, min.peaks = 0)
# Testing for differential transcript usage
res.table = DUTest(peaks.seurat,
population.1 = "stress",
population.2 = "fibroblast",
exp.thresh = 0.1,
feature.type = c("UTR3", "exon"))
res.table.top <- subset(res.table, abs(Log2_fold_change) > 1)
head(res.table.top)
# Plotting gene expression using the Seurat FeaturePlot function
Seurat::FeaturePlot(Low_L07, "ENG", cols = c("lightgrey", "red"))
# Select top peaks DU within the MMP3 gene
peaks.to.plot <- rownames(subset(res.table.top, gene_name == "ENG"))
PlotRelativeExpressionUMAP(peaks.seurat, peaks.to.plot = peaks.to.plot)
### Sierra coverage plots
cells.df <- Idents(Low_L07) %>% as.data.frame()
cells.df$cellbc <- rownames(cells.df)
colnames(cells.df) <- c("celltype", "cellbc")
gtf_gr <- rtracklayer::import("genes.gtf")
outdir = "bam_subsets/"
dir.create(outdir)
SplitBam(bam = "./L07/possorted_genome_bam.bam",
cellbc.df = cells.df,
outdir = outdir,
gtf_gr = gtf_gr,
geneSymbol = "ENG")
from sierra.
Hi Nelson,
If you're using CellRanger, it's possible you need a '-1' character appended to your cell barcodes.
Try the following code, and let me know if that helps:
cells.df$cellbc <- paste0(cells.df$cellbc, "-1")
Cheers,
- Ralph
from sierra.
Hi Ralph,
Thanks for your suggestion, but still "No data found". Any other solution? Thanks!
Regards,
Nelson
> cells.df$cellbc <- paste0(cells.df$cellbc, "-1")
> str(cells.df)
'data.frame': 1872 obs. of 2 variables:
$ celltype: Factor w/ 4 levels "cartilaginous",..: 1 3 3 3 3 3 1 3 3 2 ...
$ cellbc : chr "AAACCTGGTTAAGGGC-1" "AAACCTGTCGAGAGCA-1" "AAACGGGAGAACAATC-1" "AAACGGGAGAATCTCC-1" ...
> SplitBam(bam = "./L07/possorted_genome_bam.bam",
+ cellbc.df = cells.df,
+ outdir = outdir,
+ gtf_gr = gtf_gr,
+ geneSymbol = "ENG")
splitting bam file: ./L07/possorted_genome_bam.bam
[1] cartilaginous stress angiogenic fibroblast
Levels: cartilaginous angiogenic stress fibroblast
processing cell type cartilaginous
[W::hts_idx_load2] The index file is older than the data file: ./L07/possorted_genome_bam.bam.bai
chunk0: 8192 length of aln: 0
Writing to bam_subsets/cartilaginous.ENG.bam
processing cell type stress
[W::hts_idx_load2] The index file is older than the data file: ./L07/possorted_genome_bam.bam.bai
chunk0: 8192 length of aln: 0
Writing to bam_subsets/stress.ENG.bam
processing cell type angiogenic
[W::hts_idx_load2] The index file is older than the data file: ./L07/possorted_genome_bam.bam.bai
chunk0: 8192 length of aln: 0
Writing to bam_subsets/angiogenic.ENG.bam
processing cell type fibroblast
[W::hts_idx_load2] The index file is older than the data file: ./L07/possorted_genome_bam.bam.bai
chunk0: 8192 length of aln: 0
Writing to bam_subsets/fibroblast.ENG.bam
> sessionInfo()
R version 3.6.1 (2019-07-05)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Ubuntu 18.04.3 LTS
Matrix products: default
BLAS: /usr/lib/x86_64-linux-gnu/blas/libblas.so.3.7.1
LAPACK: /usr/lib/x86_64-linux-gnu/lapack/liblapack.so.3.7.1
locale:
[1] LC_CTYPE=en_HK.UTF-8 LC_NUMERIC=C LC_TIME=en_HK.UTF-8 LC_COLLATE=en_HK.UTF-8 LC_MONETARY=en_HK.UTF-8
[6] LC_MESSAGES=en_HK.UTF-8 LC_PAPER=en_HK.UTF-8 LC_NAME=C LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_HK.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] dplyr_0.8.3 stringr_1.4.0 Seurat_3.1.1 Sierra_0.1.0
loaded via a namespace (and not attached):
[1] reticulate_1.13 R.utils_2.9.0 tidyselect_0.2.5 RSQLite_2.1.3
[5] AnnotationDbi_1.48.0 htmlwidgets_1.5.1 grid_3.6.1 BiocParallel_1.20.0
[9] Rtsne_0.15 munsell_0.5.0 codetools_0.2-16 ica_1.0-2
[13] statmod_1.4.32 future_1.15.1 colorspace_1.4-1 Biobase_2.46.0
[17] knitr_1.26 rstudioapi_0.10 stats4_3.6.1 SingleCellExperiment_1.8.0
[21] ROCR_1.0-7 gbRd_0.4-11 listenv_0.7.0 labeling_0.3
[25] Rdpack_0.11-0 GenomeInfoDbData_1.2.2 hwriter_1.3.2 bit64_0.9-7
[29] farver_2.0.1 vctrs_0.2.0 xfun_0.11 biovizBase_1.34.0
[33] BiocFileCache_1.10.2 R6_2.4.1 GenomeInfoDb_1.22.0 rsvd_1.0.2
[37] locfit_1.5-9.1 AnnotationFilter_1.10.0 bitops_1.0-6 DelayedArray_0.12.0
[41] assertthat_0.2.1 SDMTools_1.1-221.2 scales_1.1.0 nnet_7.3-12
[45] gtable_0.3.0 npsurv_0.4-0 globals_0.12.4 ensembldb_2.10.2
[49] rlang_0.4.2 zeallot_0.1.0 genefilter_1.68.0 splines_3.6.1
[53] rtracklayer_1.46.0 lazyeval_0.2.2 acepack_1.4.1 dichromat_2.0-0
[57] checkmate_1.9.4 reshape2_1.4.3 GenomicFeatures_1.38.0 backports_1.1.5
[61] Hmisc_4.3-0 tools_3.6.1 ggplot2_3.2.1 gplots_3.0.1.1
[65] RColorBrewer_1.1-2 BiocGenerics_0.32.0 ggridges_0.5.1 Rcpp_1.0.3
[69] plyr_1.8.4 base64enc_0.1-3 progress_1.2.2 zlibbioc_1.32.0
[73] purrr_0.3.3 RCurl_1.95-4.12 prettyunits_1.0.2 rpart_4.1-15
[77] openssl_1.4.1 pbapply_1.4-2 cowplot_1.0.0 S4Vectors_0.24.1
[81] zoo_1.8-6 SummarizedExperiment_1.16.0 ggrepel_0.8.1 cluster_2.1.0
[85] magrittr_1.5 data.table_1.12.6 lmtest_0.9-37 RANN_2.6.1
[89] ProtGenerics_1.18.0 fitdistrplus_1.0-14 matrixStats_0.55.0 hms_0.5.2
[93] lsei_1.2-0 xtable_1.8-4 XML_3.98-1.20 IRanges_2.20.1
[97] gridExtra_2.3 compiler_3.6.1 biomaRt_2.42.0 tibble_2.1.3
[101] KernSmooth_2.23-16 crayon_1.3.4 R.oo_1.23.0 htmltools_0.4.0
[105] Formula_1.2-3 tidyr_1.0.0 geneplotter_1.64.0 RcppParallel_4.4.4
[109] DBI_1.0.0 dbplyr_1.4.2 MASS_7.3-51.4 rappdirs_0.3.1
[113] Matrix_1.2-18 R.methodsS3_1.7.1 gdata_2.18.0 parallel_3.6.1
[117] Gviz_1.30.0 metap_1.1 igraph_1.2.4.2 GenomicRanges_1.38.0
[121] pkgconfig_2.0.3 GenomicAlignments_1.22.1 foreign_0.8-72 plotly_4.9.1
[125] foreach_1.4.7 annotate_1.64.0 XVector_0.26.0 bibtex_0.4.2
[129] DEXSeq_1.32.0 VariantAnnotation_1.32.0 BSgenome.Hsapiens.UCSC.hg38_1.4.1 digest_0.6.23
[133] sctransform_0.2.0 RcppAnnoy_0.0.14 tsne_0.1-3 Biostrings_2.54.0
[137] leiden_0.3.1 htmlTable_1.13.2 uwot_0.1.4 curl_4.3
[141] Rsamtools_2.2.1 gtools_3.8.1 lifecycle_0.1.0 nlme_3.1-142
[145] jsonlite_1.6 viridisLite_0.3.0 askpass_1.1 BSgenome_1.54.0
[149] pillar_1.4.2 lattice_0.20-38 httr_1.4.1 survival_2.44-1.1
[153] glue_1.3.1 png_0.1-7 iterators_1.0.12 bit_1.1-14
[157] stringi_1.4.3 blob_1.2.0 DESeq2_1.26.0 latticeExtra_0.6-28
[161] caTools_1.17.1.3 memoise_1.1.0 irlba_2.3.3 future.apply_1.3.0
[165] ape_5.3
from sierra.
Hi Nelson,
I don't see the 'no data found' message in your output after the cell barcodes were updated - the messages indicate the BAM files should have been written. Can you clarify if the BAM files have not been produced in your output directory?
Cheers,
- Ralph
from sierra.
Hi Ralph,
Sorry. I check the output dir and get the bam files. It did solve the problem.
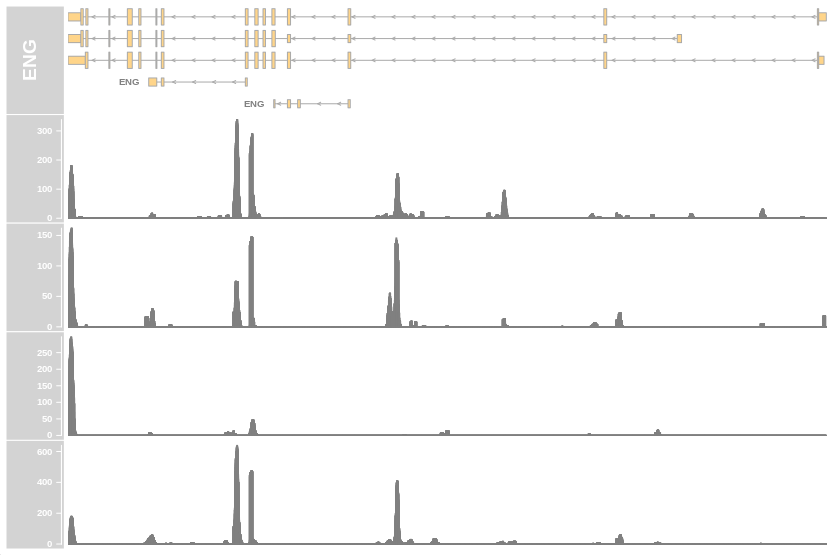
Thanks you so much for your kindly help. I love the coverage plot.
Regards,
Nelson
bam.files <- paste0(outdir, dir(outdir, "ENG.bam$"))
PlotCoverage(genome_gr = gtf_gr,
geneSymbol = "ENG",
genome = "mm10",
bamfiles = bam.files)
from sierra.
Related Issues (20)
- Using data with batch effects in Sierra HOT 2
- Error in peak calling HOT 2
- peak discrepancy HOT 1
- Error when running CountPeaks HOT 8
- CoveragePlot error with 'zoom_3UTR=TRUE ' HOT 1
- Which alignment method and indexing options are suitable to use with Sierra? HOT 3
- Does the Sierra package have a detailed protocol, I would like to find. HOT 1
- Chromosome name in FindPeaks // Help with Output HOT 3
- Generate GitHub Releases HOT 2
- Cellranger mkref function parameters for Sierra HOT 1
- FindPeaks Error--'x' values larger than vector length 'sum(width)' HOT 2
- DUTest function Error HOT 1
- Sierra dataframe has 0 length HOT 1
- [E::hts_open_format] Failed to open file HOT 1
- is it possible to generate a plot that shows global 3'UTR length change?
- issues with generating splice junction file HOT 7
- MergePeakCoordinates takes long time! HOT 9
- Paired-end & PlotRelativeExpression functions. HOT 1
- Getting "Error in (function (x) : attempt to apply non-function" HOT 3
- Using Sierra with Singleron Biotechnologies Platform HOT 1
Recommend Projects
-
 React
React
A declarative, efficient, and flexible JavaScript library for building user interfaces.
-
Vue.js
🖖 Vue.js is a progressive, incrementally-adoptable JavaScript framework for building UI on the web.
-
 Typescript
Typescript
TypeScript is a superset of JavaScript that compiles to clean JavaScript output.
-
TensorFlow
An Open Source Machine Learning Framework for Everyone
-
Django
The Web framework for perfectionists with deadlines.
-
Laravel
A PHP framework for web artisans
-
D3
Bring data to life with SVG, Canvas and HTML. 📊📈🎉
-
Recommend Topics
-
javascript
JavaScript (JS) is a lightweight interpreted programming language with first-class functions.
-
web
Some thing interesting about web. New door for the world.
-
server
A server is a program made to process requests and deliver data to clients.
-
Machine learning
Machine learning is a way of modeling and interpreting data that allows a piece of software to respond intelligently.
-
Visualization
Some thing interesting about visualization, use data art
-
Game
Some thing interesting about game, make everyone happy.
Recommend Org
-
Facebook
We are working to build community through open source technology. NB: members must have two-factor auth.
-
Microsoft
Open source projects and samples from Microsoft.
-
Google
Google ❤️ Open Source for everyone.
-
Alibaba
Alibaba Open Source for everyone
-
D3
Data-Driven Documents codes.
-
Tencent
China tencent open source team.

from sierra.