R has excellent tools for dates and times. The Date and POSIXct classes (as well as the 'wide'
representation in POSIXlt) are versatile, and a lot of useful tooling has been built around them.
However, POSIXct is implemented as a double with fractional seconds since the epoch. Given the
53 bits accuracy, it leaves just a bit less than microsecond resolution. Furthermore, using
floating-point arithmetic for an integer concept opens the door to painful issues of error
accumulation.
More and more performance measurements, latency statistics, etc., are now measured more finely, and we
need nanosecond resolution for which commonly an integer64 is used to represent nanoseconds
since the epoch.
And while R does not have a native type for this, the bit64
package by Jens Oehlschlägel offers a performant one implemented as a
lightweight S3 class. So this package uses the integer64 class, along with multiple helper functions
for parsing and formatting at nano-second resolution from the RcppCCTZ
package which wraps the CCTZ library from Google. CCTZ is a modern C++11 library
extending the (C++11-native) chrono type.
In addition to the point-in-time type nanotime, this package also provides an interval type
nanoival which may have open or closed start/end, a period type nanoperiod that is a human
representation of time, such as day, month, etc., and a duration type nanoduration. These types
are similar to what the lubridate package proposes.
Set and arithmetic operations on these types are available. All functionality is designed to
correctly handle instances across different time zones. Because these temporal types are based on R
built-in types, most functions have an efficient implementation and the types are suitable for use
in data.frame and data.table. nanotime is also a better choice than the native POSIXct in
most of the cases where fractional seconds are needed because it avoids floating point issues.
Package documentation, help pages, a vignette, and more is available here.
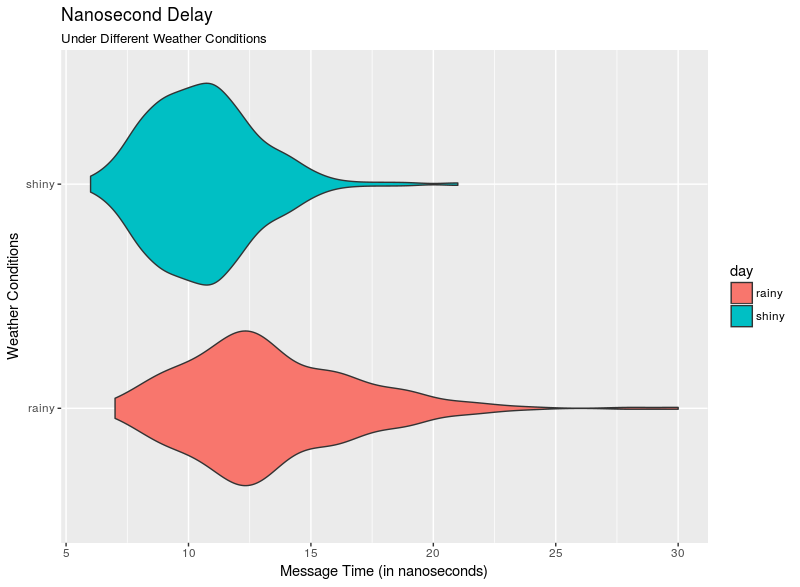
See the included demo script nanosecondDelayExample.R for a (completely simulated and hence made-up) study of network latency measured in nanoseconds resulting in the figure below
R> x <- as.nanotime("1970-01-01T00:00:00.000000001+00:00")
R> x
[1] "1970-01-01T00:00:00.000000001+00:00"
R> x + 1e9
[1] "1970-01-01T00:00:01.000000001+00:00"
R> as.nanotime("2020-03-21 Europe/London")
[1] 2020-03-21T00:00:00+00:00R> options("width"=60)
R> v <- nanotime(Sys.time()) + 1:5
R> v
[1] 2020-03-22T03:09:20.732122001+00:00
[2] 2020-03-22T03:09:20.732122002+00:00
[3] 2020-03-22T03:09:20.732122003+00:00
[4] 2020-03-22T03:09:20.732122004+00:00
[5] 2020-03-22T03:09:20.732122005+00:00
R>R> library(zoo)
R> z <- zoo(cbind(A=1:5, B=5:1), v)
R> options(nanotimeFormat="%H:%M:%E*S") ## override default format
R> z
R> options(nanotimeFormat=NULL) ## go back to default format
R> zR> library(data.table)
R> dt <- data.table(v, cbind(A=1:5, B=5:1))
R> fwrite(dt, file="datatableTest.csv") # write out
R> dtcheck <- fread("datatableTest.csv") # read back
R> dtcheck
R> dtcheck[, v:=nanotime(v)] # read as a string, need to re-class as nanotime
R> fread("../datatableTest.csv", colClasses=c("nanotime", "integer", "integer"))This requires version 0.0.2 or later.
R> data.frame(cbind(A=1:5, B=5:1), v=v)R> ival <- as.nanoival("+2009-01-01 13:12:00 America/New_York -> 2009-02-01 15:11:03 America/New_York-")
R> ival
[1] +2009-01-01T18:12:00+00:00 -> 2009-02-01T20:11:03+00:00-
R> start <- nanotime("2009-01-01 13:12:00 America/New_York")
R> end <- nanotime("2009-02-01 15:11:00 America/New_York")
R> nanoival(start, end) # by default sopen=F,eopen=T
[1] +2009-01-01T18:12:00+00:00 -> 2009-02-01T20:11:00+00:00-
R> nanoival(start, end, sopen=FALSE, eopen=TRUE)
[1] +2009-01-01T18:12:00+00:00 -> 2009-02-01T20:11:00+00:00-
R> intersect(as.nanoival("+2019-03-01 UTC -> 2020-03-01 UTC-"),
as.nanoival("+2020-01-01 UTC -> 2020-06-01 UTC-"))
[1] +2020-01-01T00:00:00+00:00 -> 2020-03-01T00:00:00+00:00-
R> union(as.nanoival("+2019-03-01 UTC -> 2020-03-01 UTC-"),
as.nanoival("+2020-01-01 UTC -> 2020-06-01 UTC-"))
[1] +2019-03-01T00:00:00+00:00 -> 2020-06-01T00:00:00+00:00-
R> setdiff(as.nanoival("+2019-03-01 UTC -> 2020-03-01 UTC-"),
as.nanoival("+2020-01-01 UTC -> 2020-06-01 UTC-"))
[1] +2019-03-01T00:00:00+00:00 -> 2020-01-01T00:00:00+00:00- R> as.nanoperiod("1y1m1w1d/01:01:01.000_000_001")
[1] 13m8d/01:01:01.000_000_001
R> nanoperiod(months=13, days=-1, duration="01:00:00")
[1] 13m-1d/01:00:00
R> ones <- as.nanoperiod("1y1m1w1d/01:01:01.000_000_001")
R> nanoperiod.month(ones); nanoperiod.day(ones); nanoperiod.nanoduration(ones)
[1] 13
[1] 8
[1] 01:01:01.000_000_001
R> plus(v, as.nanoperiod("1y1m"), tz="UTC")
[1] 2021-04-22T03:09:20.732122001+00:00
[2] 2021-04-22T03:09:20.732122002+00:00
[3] 2021-04-22T03:09:20.732122003+00:00
[4] 2021-04-22T03:09:20.732122004+00:00
[5] 2021-04-22T03:09:20.732122005+00:00R> nanoduration(hours=1, minutes=1, seconds=1, nanoseconds=1)
R> as.nanoduration("00:00:01")
R> as.nanoduration("-00:00:01")
R> as.nanoduration("100:00:00")
R> as.nanoduration("00:00:00.000_000_001")
R> from <- as.nanotime("2018-09-14T12:44:00+00:00")
R> seq(from, by=as.nanoperiod("1y"), length.out=4, tz="Europe/London")
[1] 2018-09-14T12:44:00+00:00
[2] 2019-09-14T12:44:00+00:00
[3] 2020-09-14T12:44:00+00:00
[4] 2021-09-14T12:44:00+00:00
The bit64 package (by Jens
Oehlschlägel) supplies the integer64 type used to store the nanosecond
resolution time as (positive or negative) offsets to the epoch of January 1, 1970. The
RcppCCTZ package supplies the formatting and
parsing routines based on the (modern C++) library CCTZ from
Google, when the parsing cannot be done using a fast built-in parser. integer64 is also used for
the type nanoduration, whereas nanoival and nanoperiod are stored in a complex, i.e. over
128 bits.
The package is by now fairly mature, has been rewritten once (to go from S3 to S4) and has recently received a sizeable feature extension. There may still be changes, though there should generally never be breaking ones. The package also has an extensive test suite, and very good code coverage.
See the issue tickets for an up to date list of potentially desirable, possibly planned, or at least discussed items.
The package is on CRAN and can be installed via a standard
install.packages("nanotime")whereas in order to install development versions a
remotes::install_github("eddelbuettel/nanotime") # dev versionshould suffice.
Dirk Eddelbuettel and Leonardo Silvestri
GPL (>= 2)