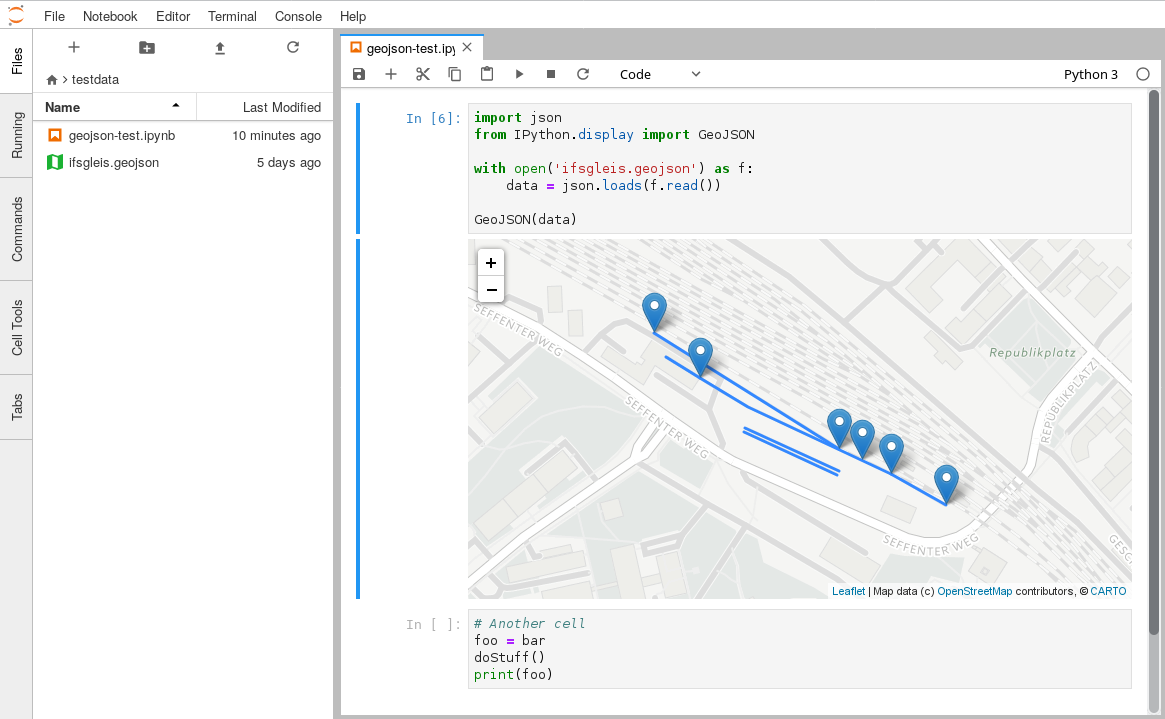
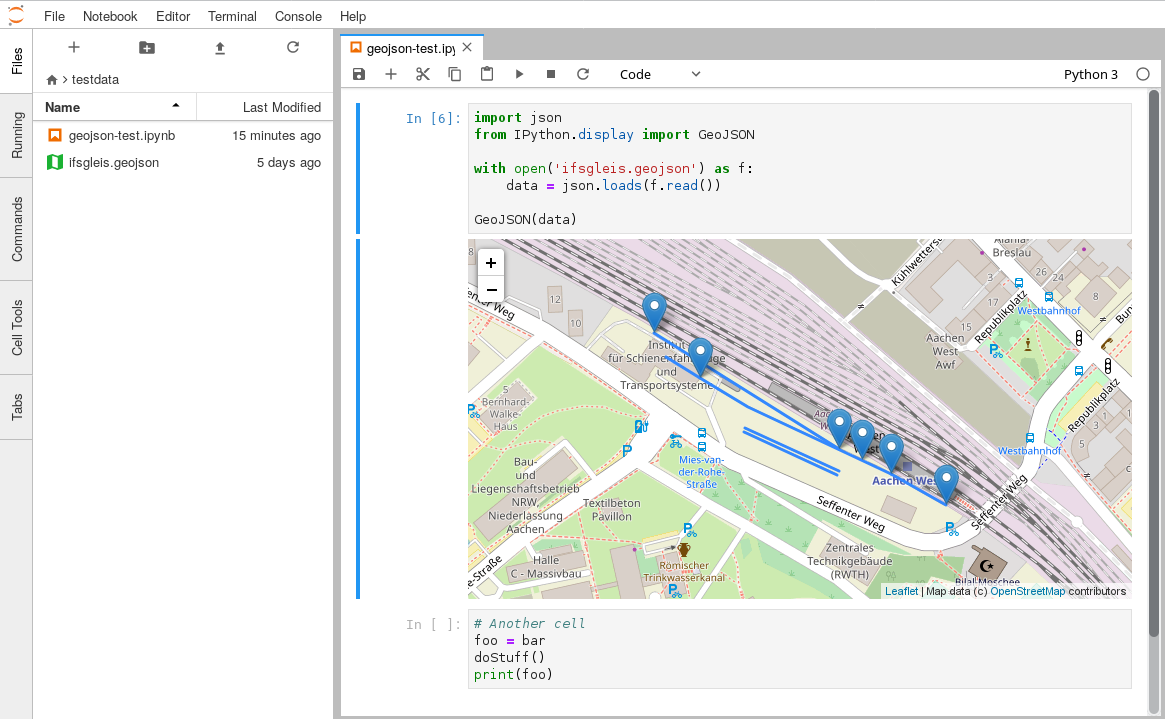
This is a monorepo that consists of JupyterLab mimerender extensions for common file and MIME types.
| Name | Mime types | File extensions | Info |
|---|---|---|---|
| fasta-extension | application/vnd.fasta.fasta |
.fasta |
 |
| geojson-extension | application/geo+json |
.geojson, .geo.json |
 |
| katex-extension | N/A | N/A |  |
| mathjax2-extension | N/A | N/A |  |
| vega3-extension | application/vnd.vega.v3+json, application/vnd.vegalite.v2+json |
.vg, .vl, .vg.json, .vl.json, .vega, .vegalite |
 |
@jupyterlab/plotly-extension is being deprecated. Please use the Plotly-supported jupyterlab-plotly. See the plotly.py README for more info.
With JupyterLab 3.0 and above, it is possible to install the prebuilt extensions with pip:
pip install jupyterlab-fasta
pip install jupyterlab-geojson
pip install jupyterlab-katex
pip install jupyterlab-mathjax2
pip install jupyterlab-vega3If you would like to contribute to the project, please read our contributor documentation.
JupyterLab follows the official Jupyter Code of Conduct.
- Node.js >= 18 (see Installing Node.js and jlpm in the JupyterLab docs)
The jlpm command is JupyterLab's pinned version of
yarn that is installed with JupyterLab. You may use
yarn or npm in lieu of jlpm below.
# Clone the repo to your local environment
git clone https://github.com/jupyterlab/jupyter-renderers.git
cd jupyter-renderers
# install the fasta extension
cd packages/fasta-extension
# Install package in development mode
pip install -e .
# Link your development version of the extension with JupyterLab
jupyter labextension develop . --overwrite
# Rebuild the extensions TypeScript source after making changes
jlpm run buildAfter making changes to the source packages, the packages must be rebuilt:
# Rebuild all the extensions at once
jlpm run build
# To rebuilt a particular extension, for example the fasta extension
cd packages/fasta-extension
jlpm run buildYou may also watch a particular extension directory for changes and automatically rebuild:
# In one terminal tab, watch the jupyter-fasta directory
cd packages/fasta-extension
jlpm run watch
# Run JupyterLab in another terminal
jupyter labBuild all Python packages:
rm -rf dist/*
jlpm build-py
twine upload dist/jupyterlab*
To create a JupyterLab icon for a new MIME or file type, you can use the Sketch file in this repo or fork the file on Figma and export your icon as an SVG. See the GeoJSON icon for reference.