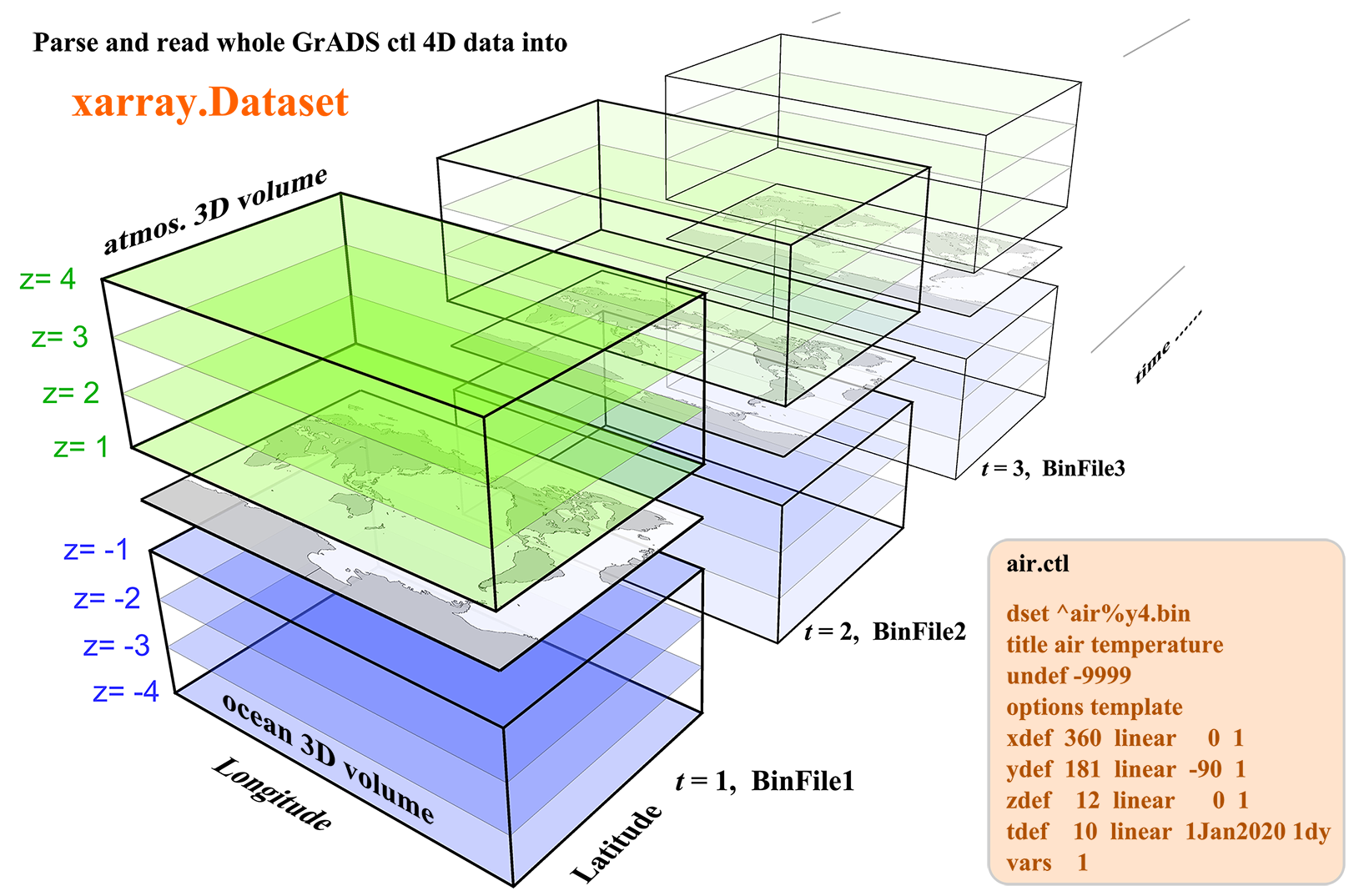
The Grid Analysis and Display System (GrADS or OpenGrADS) is a widely used software for easy access, manipulation, and visualization of earth science data. It uses a descriptor (or control) file with a suffix .ctl to describe a raw binary 4D dataset. The ctl file is similar to the header information of a NetCDF file, containing all the information about dimensions, attributes, and variables except for the variable data.
This python package xgrads is designed for parse and read the .ctl file commonly used by GrADS. Right now it can parse various kinds of .ctl files. However, only the commonly used raw binary 4D datasets can be read using dask and return as a xarray.Dataset Other types of binary data, like dtype is station orgrib, may be supported in the future.
Requirements
xgrads is developed under the environment with xarray (=version 0.15.0), dask (=version 2.11.0), numpy (=version 1.15.4), cartopy (=version 0.17.0), and pyproj (=version 1.9.6). Older versions of these packages are not well tested.
Install via pip
pip install xgrads
Install from github
git clone https://github.com/miniufo/xgrads.git
cd xgrads
python setup.py install
Parsing a .ctl file is pretty simple using the following code:
from xgrads import CtlDescriptor
ctl = CtlDescriptor(file='test.ctl')
# print all the info in ctl file
print(ctl)If you have already load the ASCII content in the .ctl file, you can do it as:
content = \
"dset ^binary.dat\n" \
"* this is a comment line\n" \
"title 10-deg resolution model\n" \
"undef -9.99e8\n" \
"xdef 36 linear 0 10\n" \
"ydef 19 linear -90 10\n" \
"zdef 1 linear 0 1\n" \
"tdef 1 linear 00z01Jan2000 1dy\n" \
"vars 1\n" \
"test 1 99 test variable\n" \
"endvars\n"
ctl = CtlDescriptor(content=content)
# print all the info
print(ctl)Reading a .ctl related binary data file is also pretty simple using the following code:
from xgrads import open_CtlDataset
dset = open_CtlDataset('test.ctl')
# print all the info in ctl file
print(dset)Then you have the dset as a xarray.Dataset. This is similar to xarray.open_dataset that use dask to chunk (buffer) parts of the whole dataset in physical memory if the whole dataset is too large to fit in.
If there are many .ctl files in a folder, we can also open all of them in a single call of open_mfdataset as:
from xgrads import open_mfDataset
dset = open_mfDataset('./folder/*.ctl')
# print all the info in ctl file
print(dset)assuming that every .ctl file has similar data structure except the time step is different. This is similar to xarray.open_mfdataset.
With the above functionality, it is easy to convert a .ctl (GrADS) dataset to a .nc (NetCDF) dataset:
from xgrads import open_CtlDataset
open_CtlDataset('input.ctl').to_netcdf('output.nc')