TL;DR: DeepMind a company affiliated with Google and specialized in AI presented a novel algorithm for Protein Structure Prediction at CASP13 (a competition which goal is to find the best algorithms that predict protein structures in different categories).
The Protein Folding Problem is an interesting one since there's tons of DNA sequence data available and it's becoming cheaper and cheaper at an unprecedented rate (faster than Moore's law). The cells build the proteins they need through transcription (from DNA to RNA) and translation (from RNA to Aminocids (AAs)). However, the function of a protein does not depend solely on the sequence of AAs that form it, but also their spatial 3D folding. Thus, it's hard to predict the function of a protein from its DNA sequence. AI can help solve this problem by learning the relations that exist between a determined sequence and its spatial 3D folding.
The DeepMind work presented @ CASP was not a technological breakthrough (they did not invent any new type of AI) but an engineering one: they applied well-known AI algorithms to a problem along with lots of data and computing power and found a great solution through model design, feature engineering, model ensembling and so on. DeepMind has no plan to open source the code of their model nor set up a prediction server.
Based on the premise exposed before, the aim of this project is to build a model suitable for protein 3D structure prediction inspired by AlphaFold and many other AI solutions that may appear and achieve SOTA results.
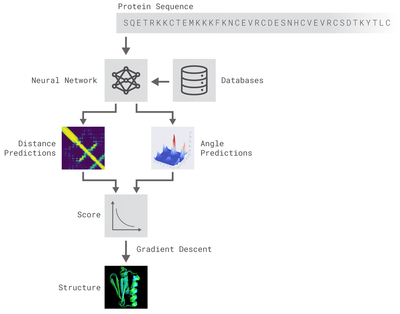
The methods implemented are inspired by DeepMind's original post. Two different residual neural networks (ResNets) are used to predict angles between adjacent aminoacids (AAs) and distance between every pair of AAs of a protein. For distance prediction a 2D Resnet was used while for angles prediction a 1D Resnet was used.
Image from DeepMind's original blogpost.
Implementation_details can be found here together with a detailed explanation. A sample result of the distance predictor:
Ground truth (left) and predicted distances (right) by our model (the yellow squares are a lack of exact position of the AAs described in the ProteinNet dataset documentation).The future directions of the project as well as planned/work-in-progress improvements are extensively exposed in the future.md file. In a brief way, some promising ideas:
- Train with crops of 64x64, not windows of 200x200 (and average at prediction time).
- Use data from Multiple Sequence Alignments (MSA) such as paired changes bewteen AAs.
- Use distance map as potential input for angle prediction (or vice versa?) .
- Train with more data (in the cloud?)
- ...
"Science is a Work In Progress."
This project has been developed in one week by 1 person and,, therefore, many limitations have appeared. They will be listed below in order to give a sense about what this project is and what it's not.
- No usage of Multiple Sequence Alignments (MSA): The methods developed in this project don't use MSA nor MSA-based features as input.
- Computing power/memory: Development of the project has taken part in a computer with the following specs: Intel i7-6700k, 8gb RAM, NVIDIA GTX-1060Ti 6gb and 256gb of storage. The capacity for data exploration, processing, training and evaluating the models is limited.
- GPU/TPUs for training: The models were trained and evaluated on a single GPU. No cloud servers were used.
- Time: One week of development during spare time. Ideas that might be worth testing in the future are described here.
- Domain expertise: No experts in the field. The author knows the basics of Biochemistry and Deep Learnning.
- Data: The average paper about Protein Structure Prediction uses a personalized dataset acquired from the Protein Data Bank (PDB). No such dataset was used. Instead, we used a subset of the ProteinNet dataset from CASP7. Our models are trained with just 150 proteins (distance prediction) and 600 proteins (angles prediction) due to memory constraints.
Due to these limitations and/or constraints, the precission/accuracy the methods here developed can achieve is limited when compared against SOTA algorithms.
- DeepMind original blog post
- AlphaFold @ CASP13: “What just happened?”
- Siraj Raval's YT video on AlphaFold
- ProteinNet dataset
Hey there! New ideas are welcome: open/close issues, fork the repo and share your code with a Pull Request. Clone this project to your computer:
git clone https://github.com/EricAlcaide/MiniFold
By participating in this project, you agree to abide by the thoughtbot code of conduct
- Author's GitHub Profile: Eric Alcaide
- Twitter: @eric_alcaide
- Email: [email protected]