GMHCC: High-throughput Analysis of Biomolecular Data using Graph-based Multiple Hierarchical Consensus Clustering
Thanks to the development of high-throughput sequencing technologies, massive amounts of various biomolecular data have been accumulated to revolutionize the study of genomics and molecular biology. One of the main challenges in analyzing these biomolecular data is to cluster their subtypes into subpopulations to facilitate subsequent downstream analysis. Recently, many clustering methods have been developed to address those biomolecular data. However, those computational methods often suffer from many limitations such as high dimensionality, data heterogeneity and noise. Moreover, it is hardly to design a computational algorithm that can simultaneously perform well on different types of biomolecular data. Therefore, in our study, we develop a novel Graph-based Multiple Hierarchical Consensus Clustering (GMHCC) with an unsupervised graph-based feature ranking and the graph-based linking method to explore the multiple hierarchical information of the underlying partitions of the consensus clustering for multiple types of biomolecular data. Indeed, we first propose to use a graph-based unsupervised feature ranking model to measure each feature by building a graph over pairwise features and then providing each feature with a ranking. Subsequently, to maintain the diversity and robustness of basic partitions, we propose multiple diverse feature subsets to generate several basic partitions and then explore the hierarchical structures of the multiple basic partitions by refining the global consensus function. Finally, we develop a new graph-based linking method, which explicitly considers the relationships between clusters to generate the final partition. Experiments on multiple types of biomolecular data including thirty-five cancer gene expression datasets and eight single-cell RNA-seq datasets validate the effectiveness of our method over several state-of-the-art consensus clustering approaches. Furthermore, differential gene analysis, gene ontology enrichment analysis, and KEGG pathway analysis are conducted, which provides novel insights into cell developmental lineages and characterization mechanisms.
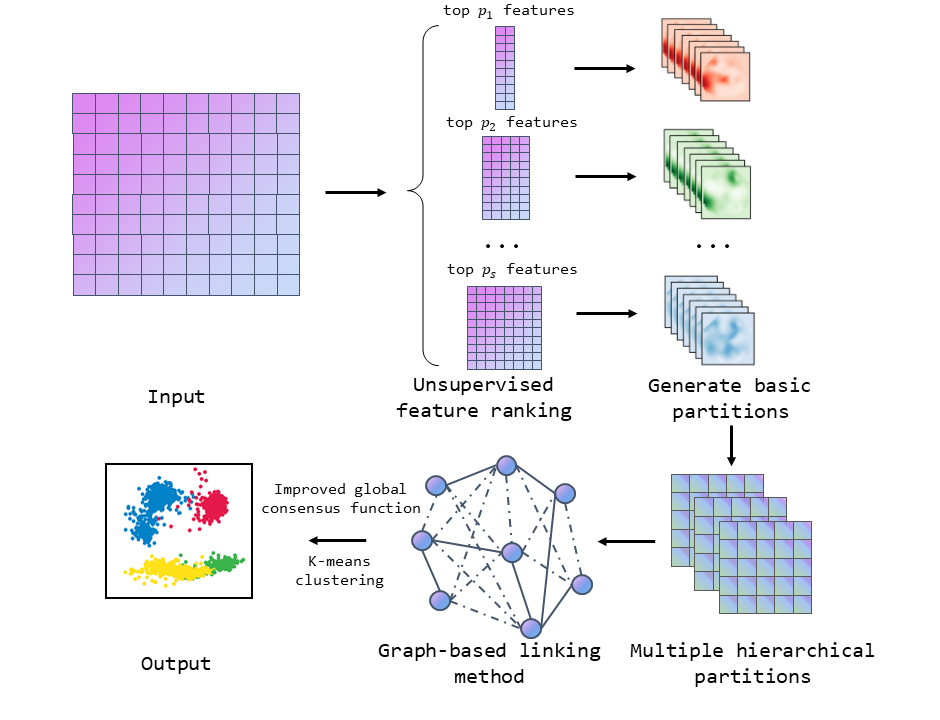
The framework of the GMHCC method. Unsupervised feature ranking method is firstly applied to input data matrix to give a ranking to features. Then a few Basic Partitions (BPs) with different proportion of selected features are integrated to multiple hierarchical basic partitions, the graph-based linking method is used to the binary consensus information matrix and produce a new graph-based information matrix. Finally, K-means clustering algorithm is applied to new information matrix and generate the final clustering result.
Our proposed GMHCC is implemented using Matlab R2016b with the following configurations: CPU: Intel(R) Core(R) I7-10700K @ 3.80GHz, RAM: 16.0GB.
The code repository for GMHCC.
The proposed GMHCC was implemented using Matlab R2016b for our paper below, and the datesets are also available in our home page. If you wang to run our codes, please download the GMHCC-master, and run GMHCC.m in Matlab. Both the software and the supporting data can be downloaded from: https://figshare.com/articles/software/GMHCC/17111291.
Lu, Y., Yu, Z., Wang, Y., Ma, Z., Wong, K. C., & Li, X. (2022). GMHCC: High-throughput Analysis of Biomolecular Data using Graph-based Multiple Hierarchical Consensus Clustering. Bioinformatics.
https://academic.oup.com/bioinformatics/advance-article/doi/10.1093/bioinformatics/btac290/6572336
The key functions of the source code and their detailed description.
| Function | Description |
|---|---|
| GMHCC.m | Main function of our method. |
| InfFS_U.m | Unsupervised Graph-based Feature Ranking |
| ComputeGM.m | Computing the graph-based linking matrix |
| Preprocess.m | Generating basic partitions |
| exMeasure.m | Computing NMI and ARI values after clustering |
For questions, comments and concerns, please contact Yifu Lu ([email protected]).