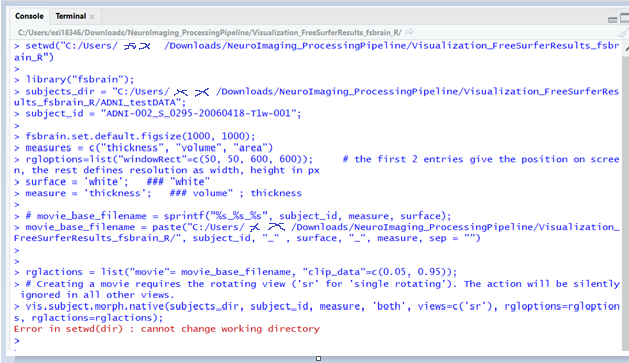
Describe the bug
The fabrain docker installation is succeed.
Within the Docker R console, I would like to create a GIF file using the demo code as below
library("fsbrain");
subjects_dir = "/home/input";
subject_id ='fsaverage'
rgloptions=list("windowRect"=c(50, 50, 600, 600));
surface = 'white';
measure = 'thickness';
movie_base_filename = paste("/home/output/", subject_id, "" , surface, "", measure, sep = "")
rglactions = list("movie"=movie_base_filename, "clip_data"=c(0.05, 0.95));
vis.subject.morph.native(subjects_dir, subject_id, measure, 'both', views=c('sr'), rgloptions=rgloptions, rglactions=rglactions);
To Reproduce
While the code is running, I can see some images are saved in the local folder.
However, the Docker R console is frozen at 200 images; showing message as "Writing '/home/output/fsaverage_white_thickness200.png' "
Then all the saved images in the local folder are all disappeared.
Then some message shows in the Docker R console as below:
Possible actions:
1: abort (with core dump, if enabled)
2: normal R exit
3: exit R without saving workspace
4: exit R saving workspace
Selection:
Selection:
Selection:
Expected behavior
A clear and concise description of what you expected to happen.
Screenshots

Environment:
- Operating System :
NAME="Ubuntu"
VERSION="18.04.5 LTS (Bionic Beaver)"
ID=ubuntu
ID_LIKE=debian
PRETTY_NAME="Ubuntu 18.04.5 LTS"
VERSION_ID="18.04"
Here I use the fsbrain docker dfspspirit/fsbrain:0.4.3
- R version (output of
R --version in system shell): R version 3.6.3 (2020-02-29)
- fsbrain version (output of
packageVersion('fsbrain') in R): 0.4.3
Looking forward to your reply and thank you very much.
Xin