Comments (54)
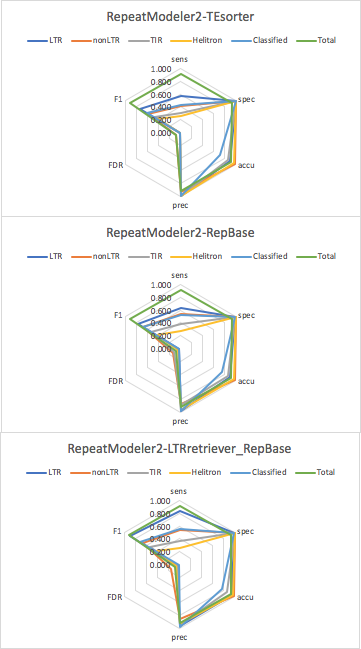
OK, I finally get the chance to run through all the pipelines associated with RepeatModeler2. The conda version RM2 v2.0.1 does not have the required RepBase library for classification, so I used TEsorter in conjunction of RM2 for Test 1. Then I also manually installed the GitHub version RM2 v2.0.1 and let RepeatClassifier do the classification for Test 2. RepeatClassifier is marginally more sensitive than TEsorter given the large database it used, but the overall specificity, accuracy, precision, and FDR are comparable among the two methods. RepBase requires license-purchasing, so TEsorter would be a free-alternative and it's much much faster.
One of the biggest improvements of RM2 is the inclusion of LTR_retriever for accurate structural identification of LTR elements. This also brings with 5 more dependencies required by the installation. The -LTRStruc module of RM2 is relatively independent and not working well with the -recoverDir function. Anyways, I got this to work after a couple of tests and shown as Test 3. Benefit from the structural module, the sensitivity of LTR annotation increased from 63.9% to 84.1%, while the annotation of other TE types is virtually unchanged.
Overall, rice TE libraries generated by RM2 had very nice annotation performance if you don't care too much about classification (i.e., Total TE annotation). The bottleneck of RM2 is in classification (license, sensitivity) and speed.
To answer the original questions, you don't need to use the -LTRStruc function for RM2 in conjunction with EDTA, because the latter incorporates LTR_retriever results as well. If you use RM2 solely, then specifying this parameter will boost the LTR annotation sensitivity for 20%.
Benchmarking plots
Raw data
| Test 1 | sens | spec | accu | prec | FDR | F1 | |
|---|---|---|---|---|---|---|---|
| LTR | RM2-TEsorter | 0.573 | 0.999 | 0.886 | 0.993 | 0.007 | 0.727 |
| nonLTR | RM2-TEsorter | 0.422 | 0.999 | 0.985 | 0.927 | 0.073 | 0.580 |
| TIR | RM2-TEsorter | 0.307 | 0.999 | 0.851 | 0.994 | 0.006 | 0.469 |
| Helitron | RM2-TEsorter | 0.261 | 1.000 | 0.966 | 0.994 | 0.006 | 0.414 |
| Classified | RM2-TEsorter | 0.436 | 0.996 | 0.705 | 0.992 | 0.008 | 0.606 |
| Total | RM2-TEsorter | 0.907 | 0.928 | 0.918 | 0.920 | 0.080 | 0.914 |
| Test 2 | sens | spec | accu | prec | FDR | F1 | |
| LTR | RM2-RepBase | 0.639 | 0.993 | 0.902 | 0.971 | 0.029 | 0.771 |
| nonLTR | RM2-RepBase | 0.547 | 0.998 | 0.987 | 0.863 | 0.137 | 0.669 |
| TIR | RM2-RepBase | 0.379 | 0.987 | 0.860 | 0.887 | 0.113 | 0.531 |
| Helitron | RM2-RepBase | 0.267 | 0.999 | 0.965 | 0.957 | 0.043 | 0.418 |
| Classified | RM2-RepBase | 0.518 | 0.989 | 0.748 | 0.980 | 0.020 | 0.678 |
| Total | RM2-RepBase | 0.907 | 0.930 | 0.919 | 0.922 | 0.078 | 0.915 |
| Test 3 | sens | spec | accu | prec | FDR | F1 | |
| LTR | RM2-LTRretriever-RepBase | 0.841 | 0.987 | 0.951 | 0.955 | 0.045 | 0.895 |
| nonLTR | RM2-LTRretriever-RepBase | 0.539 | 0.998 | 0.987 | 0.853 | 0.147 | 0.660 |
| TIR | RM2-LTRretriever-RepBase | 0.371 | 0.995 | 0.864 | 0.954 | 0.046 | 0.534 |
| Helitron | RM2-LTRretriever-RepBase | 0.262 | 0.999 | 0.965 | 0.938 | 0.062 | 0.410 |
| Classified | RM2-LTRretriever-RepBase | 0.560 | 0.990 | 0.771 | 0.982 | 0.018 | 0.713 |
| Total | RM2-LTRretriever-RepBase | 0.916 | 0.929 | 0.922 | 0.921 | 0.079 | 0.918 |
| Rice (384 Mb) | CPU allocation | Average CPU | Wall time | Max memory |
|---|---|---|---|---|
| RM2-TEsorter | 36 | 1146% | 25.5 hr | 19.0 GB |
| RM2-RepBase | 36 | ~1000% | 27.0 hr | 19.0 GB |
| RM2-LTRretriever-RepBase | 36 | 675% | 42.5 hr | 18.1 GB |
from edta.
@Juke34 OK I heard from the host of TEsorter. He said they have not yet switched to py3 and there could be maintenance issues. He recommends to put it in your github at the moment. Please let me know if you need further information.
from edta.
Yes they are, needed to use another newer genometools recipe called genometools-genometools
from edta.
A new release is needed only if the code change.
Every time you will release a new version conda will automatically update the recipe to include the last release
from edta.
from edta.
Thanks you, looking forward to have your feedback.
The EDTA recipe sounds doable I could probably help out.
from edta.
from edta.
Oh yes this is challenging, I don't know if it is possible. I think the best would be to re-implement TEsorter in python3.
from edta.
from edta.
Yes could be a solution.
I don't think we can call/imnport a container from conda.
Did you try 2to3 to see if TEsorter can be automatically converted in python3?
from edta.
from edta.
I succeeded to make TEsorter work in python3 (with pp version 1.6.4.4). Do you need drmaa for TEsorter?
from edta.
from edta.
I will probably make a repo with TEsorter for python3 and then make a conda recipe for it. But currently if I add drmaa I get this exception:
Traceback (most recent call last):
File "TEsorter.py", line 22, in <module>
from RunCmdsMP import run_cmd, pp_run
File "/Users/jacda119/git/TEsorter3/bin/RunCmdsMP.py", line 20, in <module>
import drmaa # for grid
File "//anaconda3/envs/tesorter3/lib/python3.7/site-packages/drmaa/__init__.py", line 65, in <module>
from .session import JobInfo, JobTemplate, Session
File "//anaconda3/envs/tesorter3/lib/python3.7/site-packages/drmaa/session.py", line 39, in <module>
from drmaa.helpers import (adapt_rusage, Attribute, attribute_names_iterator,
File "//anaconda3/envs/tesorter3/lib/python3.7/site-packages/drmaa/helpers.py", line 36, in <module>
from drmaa.wrappers import (drmaa_attr_names_t, drmaa_attr_values_t,
File "//anaconda3/envs/tesorter3/lib/python3.7/site-packages/drmaa/wrappers.py", line 54, in <module>
'{0}').format(_drmaa_lib_env_name))
RuntimeError: Could not find drmaa library. Please specify its full path using the environment variable DRMAA_LIBRARY_PATH
I don't know yet how to manage it, how to setup DRMAA_LIBRARY_PATH
from edta.
from edta.
In my case it crashes, I will modify the code to handle it (to no crash when missing).
from edta.
Now it is fine. I will see with the TEsorter developer to see if I push it in its repo or if I have to distribute it through my own repo.
from edta.
In my case, DRMAA_LIBRARY_PATH is a library distributed with the SGE system. Add oneline in your bashrc.
export DRMAA_LIBRARY_PATH=/software/gridengine/lib/lx-amd64/libdrmaa.sofrom edta.
I am a co-author of the TEsorter pipeline, I just sent an email to the owner and see if he likes to push the py3 version to his repo. I will let you know. Thanks for making the conversion!
from edta.
TEsorter python3 is now in bioconda.
I have changed some things, you will probably have to adjust your code to call it properly.
Once installed you can call it using TEsorter instead of TEsorter.py and now it can be run from anywhere (no need to be in the the folder as it was needed before), and no need to call the script 'build_database.sh' before.
We can start to work on a recipe to EDTA now.
from edta.
from edta.
@Juke34, I have updated EDTA to use the conda version TEsorter as well as the installation update you mentioned at the beginning of this thread. Does it fit to make a conda version EDTA now? Thanks!
from edta.
I can give a try this evening, but it is missing a test data running on a reasonable time included in the repo
from edta.
I used the Drosophila genome for testing.
Here is the genome and cds file I used:
https://de.cyverse.org/dl/d/1715CAC7-B469-41F2-B382-A9215541A770/dmel.tar.gz
This is the command I used and the final annotation result has total TE: 14.41%. The entire process took 1 hour 40 minutes.
nohup perl ~/las/git_bin/EDTA/EDTA.pl -genome dmel-all-chromosome-r6.28.fasta.rename -species others -fulllength 1 -anno 1 -evaluate 1 -t 36 -cds dmel-all-CDS-r6.30.fasta -overwrite 1 &
from edta.
I was more thinking about something running in few minutes
from edta.
You need to add a license in your repo
from edta.
@Juke34 Thanks for reminding. I just added the GPLv3 license. For quick testing, you may use a single chromosome, but the result is not representing the whole-genome level (that's fine for a run-through test).
from edta.
Could you make a new release of the tool? e.g v1.7.1
I will use it to try the conda recipe.
Then finger crossed that it will work... there are many packages... and they must no conflict and be among 3 channels :
Dependencies
There is currently no mechanism to define, in the meta.yaml file, that a particular dependency should come from a particular channel. This means that a recipe must have its dependencies in one of the following:
as-yet-unbuilt recipes in the repo but that will be included in the PR
conda-forge channel
bioconda channel
default Anaconda channel
while currently you use not available channel e.g. cyclus
from edta.
@Juke34 I will make a release for the latest version (v1.7.3) shortly and let you know. For other dependencies that are not in the three channels, is that a way to clone it to one of these three channels?
from edta.
@Juke34 EDTA v1.7.3 released!
from edta.
Conda recipe ongoing here => bioconda/bioconda-recipes#19811
from edta.
That's super exciting!!
from edta.
@Juke34 Another issue that could be resolved in the conda version of EDTA is the compilation of GRF (EDTA/bin/GenericRepeatFinder). Currently it's a precompiled version with c++11 but it often has the "GLIBC library not found" issue (#13) in other platforms. The easiest solution is to recompile it using c++11 but this is a high version and usually not available. Is it possible to recompile GRF while configuring conda? Installation of the main program should be sufficient ([1] Installation of GRF in readme).
from edta.
This has to be Fixed before to go further
from edta.
The way docker to resolve this is to recompile GRF while building the image: https://hub.docker.com/r/kapeel/edta/dockerfile. Can we do the same or similar thing? If not, what are the possible options?
from edta.
bioconda/bioconda-recipes#19841
from edta.
You are the best!!!
from edta.
I don't promise anything and t have limited time to work on that. Let's see how far we can go
from edta.
from edta.
You can remove genometools and grf from the bin for the next release and update the README.
I think you can remove --add channels biocore --add channels cyclus from the command and add genericrepeatfinder genometools-genometools in the list for the conda install command
from edta.
@Juke34 Thanks for the updates! I will test it locally and merge. Are the conflicts between TEsorter and genometools resolved?
from edta.
If you release the version 1.7.4 I should have a working recipe
from edta.
Thanks! v1.7.4 is released.
from edta.
Any particular reason you use blast-legacy and not blast?
from edta.
There was a time TIR-Learner relied on blastdbcmd and blast-legacy is the only version that has blastdbcmd behaved normally. I just double-checked neither EDTA nor TIR-Learner needs this now so it can be changed to a regular blast. Does that mean I don't even need to install blast because it has been included by other dependencies such as Repeatmasker/RepeatModeler?
from edta.
As EDTA use blast he has to be included as dependency even if it already comes with other dependencies. Conda will check and inform if there is problem with the versions required by the different dependencies and the one you need in EDTA.
Blast-legacy paths were clobbering with blast paths. Removing blast-legacy remove some warnings. And blast is more up-to-date.
from edta.
I see. Then I will just change blast-legacy to blast. Do I need to make a new release for this change?
from edta.
I don't think so it is just in the README isn't it ? The command are the same no?
from edta.
Yes, just the readme. So each time the command changes and if I want the conda version has the same update, I should make a new release right? Can the conda recipe update automatically?
from edta.
You can add ltr_retriever among the list of conda package to install in the README and remove it from bin folder too.
from edta.
I have added the ltr_retriever recipe and updated README. I am using the temporary conda repo for quick installation at the moment, and wait for updates from LTR_FINDER. I will keep you updated. I think this may be the furthest point we can reach at the moment without compromising the original usage (ie., set LTR_FINDER_parallel as optional for commercial users). I am so grateful for all your help!!!
from edta.
@Juke34 I finally get the chance to test the RM2. I am still working on it, but found some issues on the conda version:
The first issue @baozg described in here: #58 (comment)
The second issue is related to the -LTRStruc parameter. When it's specified, the program will exit with this message:
LTRPipeline dependency missing or incorrectly set for NINJA_DIR!
Rerun ./configure or check your command line to ensure that RepeatModeler
has access to and the correct version of this dependency.
Providing the path to -ninja_dir did not help, not sure if this is the issue of RM2 or the conda version.
Best,
Shujun
from edta.
For repeat classification within RepeatModeler it is done by RepeatMasker but need a library that I din't include by default in the recipe. Personally I use RepBase because my organisation pay a licence for it. By default we can include the Dfam one which is free but less good as RepBase.
Ninja2 is not yet into conda because it was not compiling on OSX, so just due to this dependency RM and EDTA would have been only Linux, it is why I was waiting for the dev to fix it. It should be done in their version 2.
from edta.
@Juke34 I used TEsorter to get around the classification of RM2 in EDTA. Outside of EDTA I also used the RepeatClassifier in RM1 installed in my institute for classification.
For NINJA I was using the old version that's the reason it wasn't recognized by RM2. After downloading the github version (NINJA2) and provide it's path to RM2 it works. I'll get back to you with the RM2 benchmarking results with and without LTR_retriever.
from edta.
Related Issues (20)
- The use of EDTA_raw.pl and EDTA.pl HOT 2
- Evaluating the results of an already finished EDTA pipeline run HOT 1
- ERROR:EDTA.pl line 583 HOT 5
- TIR-Learner does not run properly in EDTA v2.2.0 HOT 11
- LTR retriever fails HOT 5
- LINE search takes much longer compared to other steps HOT 3
- resubmit jobs HOT 1
- Rerun EDTA 2.2.0 with results from older version HOT 8
- PanEDTA Line Detection HOT 1
- 2024-01-30 02:12:48,620 -WARNING- Grid computing is not available because DRMAA not configured properly: HOT 1
- TIR filtration HOT 3
- EDTA crahed after no SINE found HOT 6
- Benefit of using panEDTA HOT 1
- Testing output HOT 3
- EDTA_2.2.x.yml seems not working HOT 7
- Concatinating CDS files for panEDTA HOT 1
- making the output gff3 valid HOT 4
- Exploring the Discrepancies between TEanno.gff3 and TElib.fa HOT 2
- Inflated TE counts and masked bp in EDTA annotation after removal of part of the genome HOT 3
- panEDTA for metazoans HOT 5
Recommend Projects
-
 React
React
A declarative, efficient, and flexible JavaScript library for building user interfaces.
-
Vue.js
🖖 Vue.js is a progressive, incrementally-adoptable JavaScript framework for building UI on the web.
-
 Typescript
Typescript
TypeScript is a superset of JavaScript that compiles to clean JavaScript output.
-
TensorFlow
An Open Source Machine Learning Framework for Everyone
-
Django
The Web framework for perfectionists with deadlines.
-
Laravel
A PHP framework for web artisans
-
D3
Bring data to life with SVG, Canvas and HTML. 📊📈🎉
-
Recommend Topics
-
javascript
JavaScript (JS) is a lightweight interpreted programming language with first-class functions.
-
web
Some thing interesting about web. New door for the world.
-
server
A server is a program made to process requests and deliver data to clients.
-
Machine learning
Machine learning is a way of modeling and interpreting data that allows a piece of software to respond intelligently.
-
Visualization
Some thing interesting about visualization, use data art
-
Game
Some thing interesting about game, make everyone happy.
Recommend Org
-
Facebook
We are working to build community through open source technology. NB: members must have two-factor auth.
-
Microsoft
Open source projects and samples from Microsoft.
-
Google
Google ❤️ Open Source for everyone.
-
Alibaba
Alibaba Open Source for everyone
-
D3
Data-Driven Documents codes.
-
Tencent
China tencent open source team.

from edta.