The official implementation for "NodeFormer: A Scalable Graph Structure Learning Transformer for Node Classification" which is accepted to NeurIPS22 as a spotlight presentation.
Related materials: [paper], [slides], [blog Chinese | English], [vedio Chinese | English], [tutorial]
Two follow-up works: DIFFormer (ICLR2023 spotlight) a physics-inspired scalable Transformer, SGFormer (NeurIPS2023) a simplified Transformer scaling to billion-scale graphs
-
[2022.11.27] We release the early version of codes for reproducibility.
-
[2023.02.20] We provide the checkpoints of NodeFormer on ogbn-Proteins and Amazon2M (see here for details).
-
[2023.03.08] We add results on cora, citeseer, pubmed with semi-supervised random splits (see here for details).
-
[2023.04.24] Another work DIFFormer (with linear attention) will appear on ICLR2023. The open source implementation is ready.
-
[2023.04.27] Upload the script for figure plotting
plot_main.ipynbwhich contains the exact scores used for our figures in the paper. -
[2023.07.03] I gave a talk on LOG seminar about scalable graph Transformers. See the online video here.
-
[2023.09.30] A follow-up work SGFormer (with simplified one-layer attentional architecture) will appear on NeurIPS2023. The open source implementation is ready.
-
[2024.03.04] Gave a 10-min talk summarizing NodeFormer, DIFFormer and SGFormer. See the video.
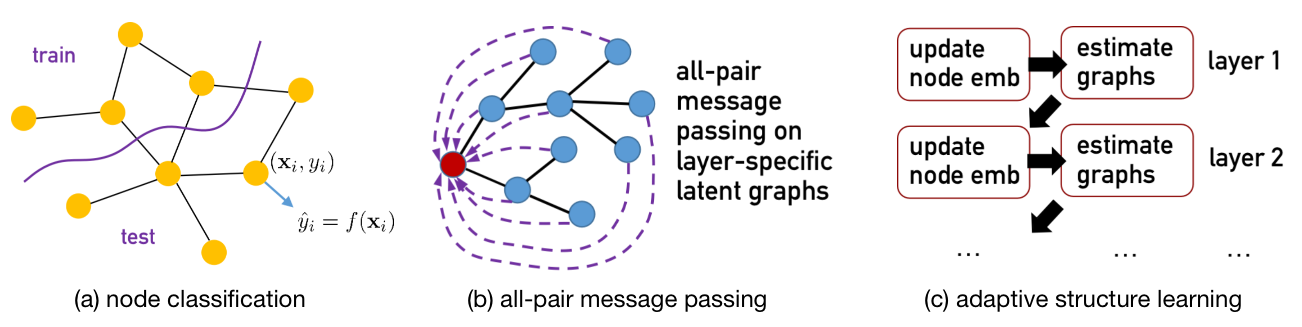
This work takes an initial step for exploring Transformer-style graph encoder networks for large node classification graphs, dubbed as NodeFormer, as an alternative to common Graph Neural Networks, in particular for encoding nodes in an input graph into embeddings in latent space.
NodeFormer is a pioneering Transformer model for node classification on large graphs. NodeFormer scales all-pair message passing with efficient latent structure learning to million-level nodes. NodeFormer has several merits:
-
All-Pair Message Passing on Layer-specific Adaptive Structures. The feature propagation per layer is operated on a latent graph that potentially connect all the nodes, in contrast with the local propagation design of GNNs that only aggregates the embeddings of neighbored nodes.
-
Linear Complexity w.r.t. Node Numbers. The all-pair message passing on latent graphs that are optimized together only requires
$O(N)$ complexity, empowered by our proposed kernelized Gumbel-Softmax operator. The largest demonstration of our model in our paper is the graph with 2M nodes, yet we believe it can even scale to larger ones with the mini-batch partition. -
Efficient End-to-End Learning for Latent Structures. The optimization for the latent structures is allowed for end-to-end training with the model, making the whole learning process simple and efficient. E.g., the training on Cora only requires 1-2 minutes, while on OGBN-Proteins requires 1-2 hours in one run.
-
Flexibility for Inductive Learning and Graph-Free Scenarios. NodeFormer is flexible for handling new unseen nodes in testing and as well as predictive tasks without input graphs, e.g., image and text classification. It can also be used for interpretability analysis with the latent interactions among data points explicitly estimated.
The key module of NodeFormer is the kernelized (Gumbel-)Softmax message passing which achieves all-pair message passing on a latent
graph in each layer with nodeformer.py implements our model:
-
The functions
kernelized_softmax()andkernelized_gumbel_softmax()implement the message passing per layer. The Gumbel version is only used for training. -
The layer class
NodeFormerConvimplements one-layer feed-forward of NodeFormer (which contains MP on a latent graph, adding relational bias and computing edge-level reg loss from input graphs if available). -
The model class
NodeFormerimplements the model that adopts standard input (node features, adjacency) and output (node prediction, edge loss).
For other files, the descriptions are below:
-
main.pyis the pipeline for full-graph training/evaluation. -
main-batch.pyis the pipeline for training with random mini-batch partition for large datasets.
We provide an easy access to the used datasets in the Google drive. This also contains other commonly used graph datasets, except the large-scale graphs OGBN-Proteins and Amazon2M which can be downloaded automatically with our codes See here for how to get the datasets ready for running our codes.
The information and sources of datasets are summarized below
-
Transductive Node Classification (Sec 4.1 in paper): we use two homophilous graphs Cora and Citeseer and two heterophilic graphs Deezer-Europe and Actor. These graph datasets are all public available at Pytorch Geometric. The Deezer dataset is provided from Non-Homophilous Benchmark, and the Actor (also called Film) dataset is provided by Geom-GCN.
-
Large Graph Datasets (Sec 4.2 in paper): we use OGBN-Proteins and Amazon2M as two large-scale datasets. These datasets are available at OGB. The original Amazon2M is collected by ClusterGCN and later used to construct the OGBN-Products.
-
Graph-Enhanced Classification (Sec 4.3 in paper): we also consider two datasets without input graphs, i.e., Mini-Imagenet and 20News-Group for image and text classification, respectively. The Mini-Imagenet dataset is provided by Matching Network, and 20News-Group is available at Scikit-Learn
| Dataset | Split | Metric | Result | Hyper-parameters/Checkpoints |
|---|---|---|---|---|
| Cora | random 50%/25%/25% | Accuracy | 88.80 (0.26) | train script |
| CiteSeer | random 50%/25%/25% | Accuracy | 76.33 (0.59) | train script |
| Deezer | random 50%/25%/25% | ROC-AUC | 71.24 (0.32) | train script |
| Actor | random 50%/25%/25% | Accuracy | 35.31 (0.89) | train script |
| OGBN-Proteins | public split | ROC-AUC | 77.45 (1.15) | train script, checkpoint, test script |
| Amazon2M | random 50%/25%/25% | Accuracy | 87.85 (0.24) | train script, checkpoint and fixed splits, test script |
| Mini-ImageNet (kNN, k=5) | random 50%/25%/25% | Accuracy | 86.77 (0.45) | train script |
| Mini-ImageNet (no graph) | random 50%/25%/25% | Accuracy | 87.46 (0.36) | train script |
| 20News-Group (kNN, k=5) | random 50%/25%/25% | Accuracy | 66.01 (1.18) | train script |
| 20News-Group (no graph) | random 50%/25%/25% | Accuracy | 64.71 (1.33) | train script |
| Cora | 20 nodes per class for train | Accuracy | 83.4 (0.2) | train script |
| CiteSeer | 20 nodes per class for train | Accuracy | 73.0 (0.3) | train script |
| Pubmed | 20 nodes per class for train | Accuracy | 81.5 (0.4) | train script |
-
Install the required package according to
requirements.txt -
Create a folder
../dataand download the datasets from here (For large graph datasets Proteins and Amazon2M, the datasets will be automatically downloaded) -
To run the training and evaluation on eight datasets we used, one can use the scripts in
run.sh:
# node classification on small datasets
python main.py --dataset cora --rand_split --metric acc --method nodeformer --lr 0.001 \
--weight_decay 5e-3 --num_layers 2 --hidden_channels 32 --num_heads 4 --rb_order 2 --rb_trans sigmoid \
--lamda 1.0 --M 30 --K 10 --use_bn --use_residual --use_gumbel --runs 5 --epochs 1000 --device 0
# node classification on large datasets
python main-batch.py --dataset ogbn-proteins --metric rocauc --method nodeformer --lr 1e-2 \
--weight_decay 0. --num_layers 3 --hidden_channels 64 --num_heads 1 --rb_order 1 --rb_trans identity \
--lamda 0.1 --M 50 --K 5 --use_bn --use_residual --use_gumbel --use_act --use_jk --batch_size 10000 \
--runs 5 --epochs 1000 --eval_step 9 --device 0
# image and text datasets
python main.py --dataset mini --metric acc --rand_split --method nodeformer --lr 0.001\
--weight_decay 5e-3 --num_layers 2 --hidden_channels 128 --num_heads 6\
--rb_order 2 --rb_trans sigmoid --lamda 1.0 --M 30 --K 10 --use_bn --use_residual --use_gumbel \
--run 5 --epochs 300 --device 0
- We also provide the checkpoints of NodeFormer on two large datasets, OGBN-Proteins and Amazon2M.
One can download the trained models into
../model/and run the scripts inrun_test_large_graph.shfor reproducing the results.
- For Amazon2M, to ensure obtaining the result as ours, one need to download the fixed splits from here to
../data/ogb/ogbn_products/split/random_0.5_0.25.
NodeFormer can in principle be applied to solve three families of tasks:
-
Node-Level Prediction on (Large) Graphs: use NodeFormer to replace GNN encoder as an encoder backbone for graph-structured data.
-
General Machine Learning Problems: use NodeFormer as an encoder that computes instance representations by their global all-pair interactions, for general ML tasks, e.g., classification or regression.
-
Learning Latent Graph Structures: use NodeFormer to learn latent graphs among a set of objects.
Our work takes an initial step for exploring how to build a scalable graph Transformer model
for node classification, and we also believe there exists ample room for further research and development
as future works. One can also use our implementation kernelized_softmax() and kernelized_gumbel_softmax()
for related projects concerning e.g., structure learning and communication, where the scalability matters.
If you find our codes useful, please consider citing our work
@inproceedings{wu2022nodeformer,
title = {NodeFormer: A Scalable Graph Structure Learning Transformer for Node Classification},
author = {Qitian Wu and Wentao Zhao and Zenan Li and David Wipf and Junchi Yan},
booktitle = {Advances in Neural Information Processing Systems (NeurIPS)},
year = {2022}
}We acknowledge the implementation of the softmax kernel
https://github.com/lucidrains/performer-pytorch
and the training pipeline for GNN node classification