VISFD is a small C++ template library for 3D image processing ("visfd.hpp") which is useful for extracting geometric shapes, volumes, and other features from 3-D volumetric images (eg. tomograms). VISFD also includes a basic C++ library for reading and writing MRC files. ("mrc_simple.hpp").
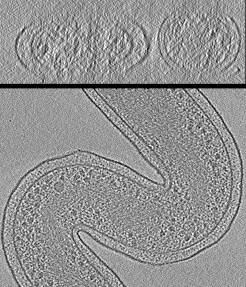
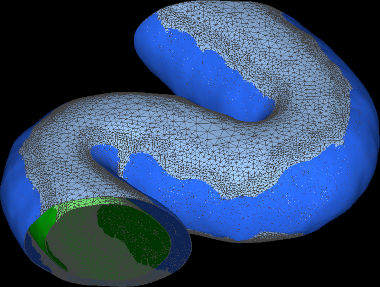
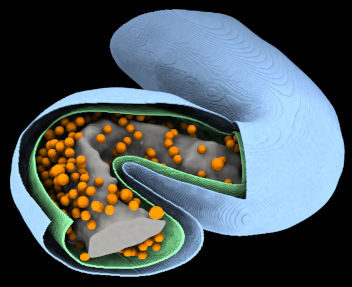
VISFD's most useful feature is probably its ability to segment cell volumes in Cryo-ET images, as well as membrane-bound organelles within cells.
VISFD is also a collection of stand-alone programs which use this library (including "filter_mrc", "combine_mrc", "voxelize_mesh", "sum_voxels", and "pval_mrc"). They are documented here. Multiprocessor support is implemented using OpenMP.
Machine-learning based detectors for membranes and large molecular complexes have been implemented in programs like EMAN2. VISFD compliments this software by providing tools for geometry extraction, curve and surface detection, signed normal determination, and robust connected-component analysis. In a typical workflow, features of interest (like membranes or filaments) would be detected and emphasized using programs like EMAN2. Then the coordinates of these geometric shapes would be extracted using VISFD and analyzed using 3rd-party tools like SSDRecon/PoissonRecon. This makes it possible to detect and close holes in incomplete membrane surfaces automatically. However, VISFD can detect curves and surfaces on it's own. (EMAN2 is recommended but not required.)
Although VISFD has similiarities and features to general libraries for for 3-D image processing, such as scikit-image and scipy.ndimage, VISFD is no where near as general or well documented as these tools. However VISFD has additional features (such as tensor-voting) which are useful for extracting geometry from Cryo-EM tomograms of living cells.
After compilation, all programs will be located in the "bin/" subdirectory. Here is a brief description of some of them:
filter_mrc is a stand-alone program which uses many of the features of the visfd library. This program was intended to be used for automatic detection and segmentation of closed membrane-bound compartments in Cryo-EM tomograms. Other features include filtering, annotation, scale-free blob-detection, morphological noise removal, connected component analysis, filament (curve) detection (planned), and edge detection (planned). Images can be segmented hierarchically into distinct contiguous objects, using a variety of strategies. This program currently only supports the .MRC (a.k.a. .REC or .MAP) image file format. As of 2021-9-13, this program does not have a graphical user interface.
Tutorials for using filter_mrc this are available here. A (long) reference manual for this program is available here. The source code for the VISFD filters used by this program is located here.
voxelize_mesh.py is a program that finds the voxels in a volumetric image that lie within the interior of a closed surface mesh. It was intended for segmenting the interiors of membrane-bound compartments in tomograms of cells. The mesh files that this program reads are typically generated by filter_mrc (together with other tools). However it can read any standard PLY file containing a closed polyhedral mesh. This program currently only supports the .mrc/.rec image file format. Documentation for this program is located here. WARNING: This experimental program is very slow and currently requires a very large amount of RAM.
combine_mrc is a program for combining two volumetric images (i.e. tomograms, both of identical size) into one image/tomogram, using a combination of addition, subtraction, multiplication, division, and thresholding operations. These features can be used perform binary operations between two images (which are similar to "and", "or", and "not" operations.) Documentation for this program is located here.
histogram_mrc.py is a graphical python program which displays the histogram of voxel intensities contained in an MRC file. It can be useful when deciding what thresholds to use with in the "filter_mrc" and "combine_mrc" programs. Voxels and regions in the image can be excluded from consideration by using the "-mask" and "-mask-select" arguments. This software requires the matplotlib and mrcfile python modules (both of which can be installed using pip). Documentation for this program is located here.
sum_voxels is a program for estimating volumes. It is a simple program which reads an MRC (.REC) file as an argument and computes the sum of all the voxel intensities. (Typically the voxel intensities are either 1 or 0. The resulting sums can be converted into volumes either by multiplying by the volume-per-voxel, or by specifying the voxel width using the "-w" argument, and including the "-volume" argument.) For convenience, threshold operation can be applied (using the "-thresh", "-thresh2", and "-thresh4" arguments) so that the voxels intensities vary between 0 and 1 before the sum is calculated. The sum can be restricted to certain regions (by using the "-mask" and "-mask-select" arguments). Documentation for this program is located here.
pval_mrc is a program for estimating the probability that a cloud of points in an image is distributed randomly. It looks for regions of high (or low) density in an image. (The user can specify a -mask argument to perform the analysis in small, confined, irregularly-shaped subvolumes from a larger image.) Documentation for this program is located here.
As of 2023, development on this software has been postponed. If I have time to work on it again, some program names, command line arguments, file names, and function names (in the API) may be altered in the future.
The INSTALL.md file has instructions for installing VISFD and its dependencies.
- 16GB of RAM or higher. (For membrane detection, your RAM must exceed 11x-44x the size of the tomogram that you are analyzing. This does not include the memory needed by the OS, browser, or other programs you are running. The voxelize_mesh.py program requires even more memory. You can reduce the memory needed and computation time dramatically by cropping or binning your tomogram.)
- A terminal (running BASH) where you can enter commands.
- A C++ compiler
- make
- Software to visualize MRC/REC/MAP files (such as IMOD/3dmod)
- python (version 3.0 or later)
- The "numpy", "matplotlib", "mrcfile", and "pyvista" python modules (These are are installable via "pip3" or "pip".)
- SSDRecon/PoissonRecon.
- Software to visualize mesh files (such as meshlab).
- A text editor. (Such as vi, emacs, atom, VisualStudio, Notepad++, ... Apple's TextEdit can be used if you save the file as plain text.)
- A computer with at least 4 CPU cores (8 threads).
- 32GB of RAM (depending on image size, this still might not be enough)
- ChimeraX is useful for visualizing MRC files in 3-D and also includes simple graphical volume editing capability.
All of the code in this repository (except for code located in "lib/mrc_simple" and "lib/visfd/eigen_simple.hpp") is available under the terms of the terms of the MIT license. (See "LICENSE.md")
The "lib/visfd/eigen3_simple.hpp" file contains code from Eigen which requires the MPL-2.0 license.
A small subset of the code in "lib/mrc_simple" was adapted from IMOD. The IMOD code uses the GPL license (version 2), which is more restrictive. License details for the "mrc_simple" library can be found in the COPYRIGHT.txt file located in that directory. If you write your own code using the "visfd" library to analyze 3D images (which you have loaded into memory by some other means), then you can ignore this notice.
VISFD was funded by NIH grant R01GM120604.
If you find this program useful, please cite DOI 10.5281/zenodo.5559243