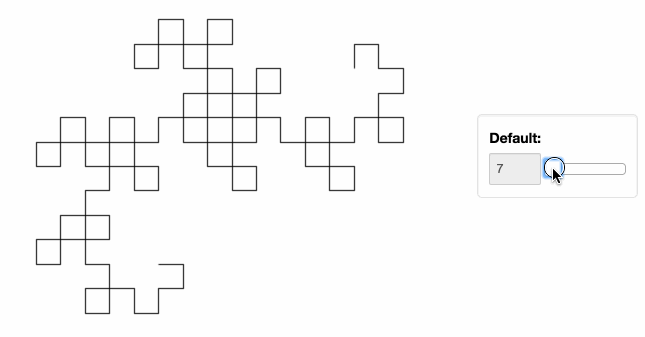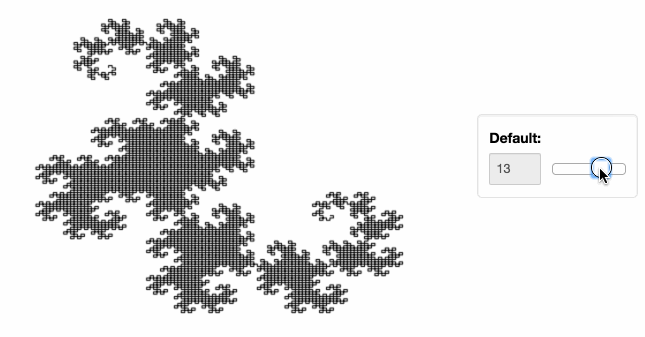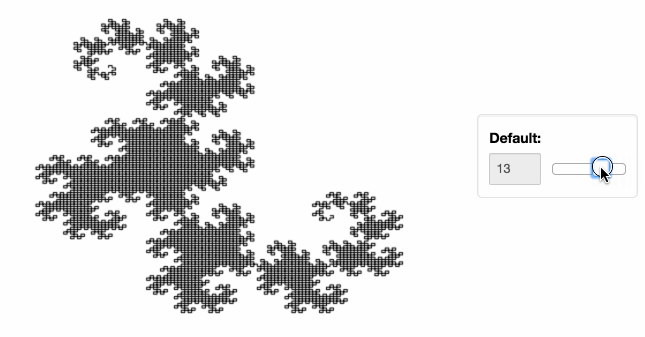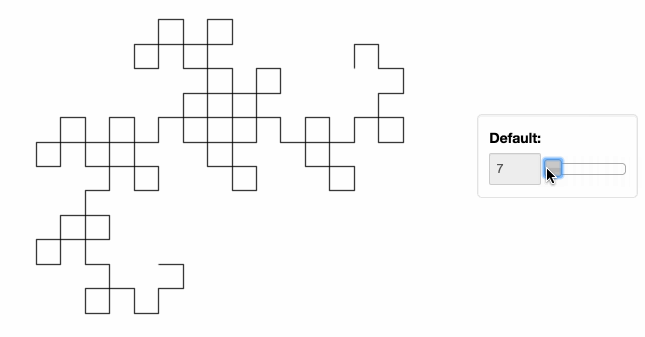
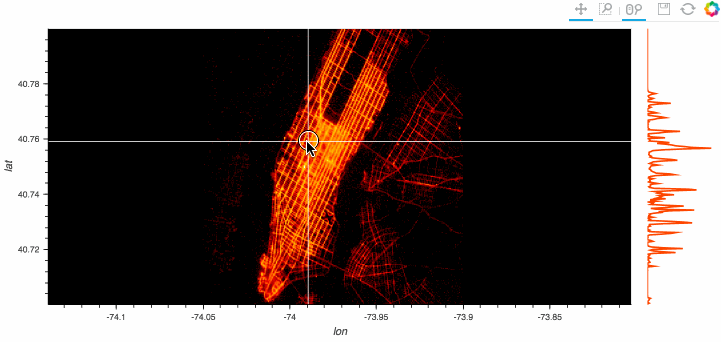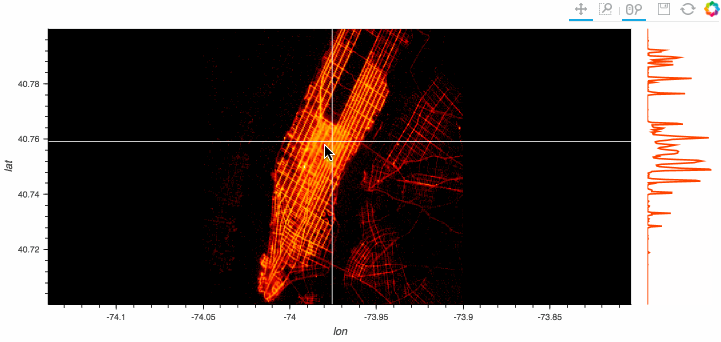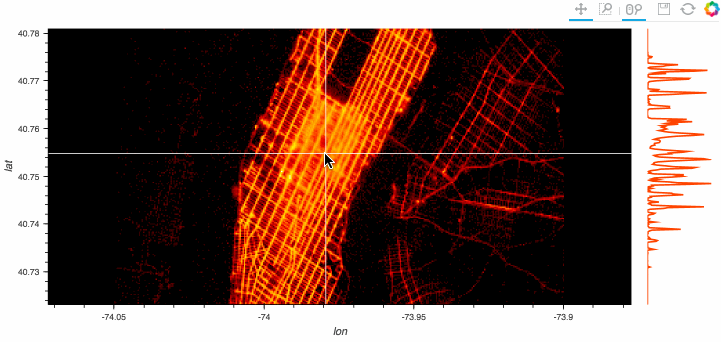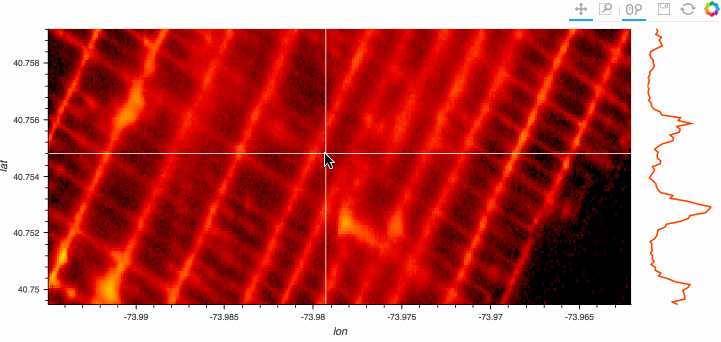
Stop plotting your data - annotate your data and let it visualize itself.
| Downloads |  |
| Build Status | |
| Coverage | |
| Latest dev release |   |
| Latest release |      |
| Python |  |
| Docs |  |
| Binder |  |
| Support |  |
HoloViews is an open-source Python library designed to make data analysis and visualization seamless and simple. With HoloViews, you can usually express what you want to do in very few lines of code, letting you focus on what you are trying to explore and convey, not on the process of plotting.
Check out the HoloViews web site for extensive examples and documentation.
HoloViews works with Python on Linux, Windows, or Mac, and works seamlessly with Jupyter Notebook and JupyterLab.
You can install HoloViews either with conda or pip, for more information see the install guide.
conda install holoviews
pip install holoviews
If you want to help develop HoloViews, you can checkout the developer guide, this guide will help you get set-up. Making it easy to contribute.
If you find any bugs or have any feature suggestions please file a GitHub issue.
If you have any usage questions, please ask them on HoloViz Discourse,
For general discussion, we have a Discord channel.