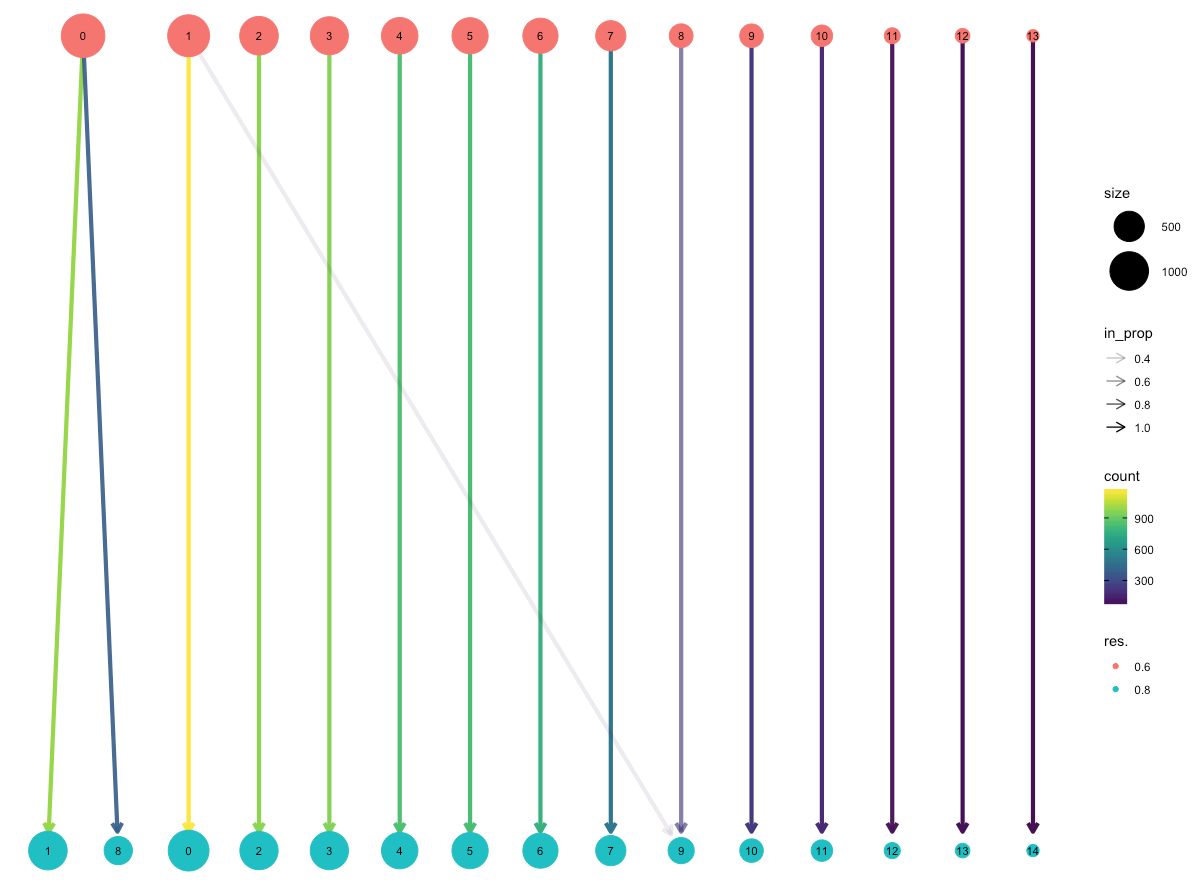
Deciding what resolution to use can be a difficult question when approaching a clustering analysis. One way to approach this problem is to look at how samples move as the number of clusters increases. This package allows you to produce clustering trees, a visualisation for interrogating clusterings as resolution increases.
You can install the release version of clustree from CRAN with:
install.packages("clustree")If you want to use the development version that can be installed from GitHub
using the remotes package:
# install.packages("remotes")
remotes::install_github("lazappi/clustree@develop")To also build the vignettes use:
# install.packages("remotes")
remotes::install_github("lazappi/clustree@develop", dependencies = TRUE,
build_vignettes = TRUE)NOTE: Building the vignettes requires the installation of additional packages.
The documentation for clustree is available from CRAN at https://cran.r-project.org/package=clustree.
To view the vignette and all the package documentation for the development version visit http://lazappi.github.io/clustree.
If you use clustree or the clustering trees approach in your work please cite our publication "Zappia L, Oshlack A. Clustering trees: a visualization for evaluating clusterings at multiple resolutions. Gigascience. 2018;7. DOI:gigascience/giy083.
citation("clustree")
Zappia L, Oshlack A. Clustering trees: a visualization for
evaluating clusterings at multiple resolutions. GigaScience.
2018;7. DOI:gigascience/giy083
A BibTeX entry for LaTeX users is
@Article{,
author = {Luke Zappia and Alicia Oshlack},
title = {Clustering trees: a visualization for evaluating clusterings at
multiple resolutions},
journal = {GigaScience},
volume = {7},
number = {7},
month = {jul},
year = {2018},
url = {http://dx.doi.org/10.1093/gigascience/giy083},
doi = {10.1093/gigascience/giy083},
}
Thank you to everyone who has contributed code to the clustree package:
- @andreamrau - added the
edge_arrow_endsoption - @mojaveazure - added support for Seurat v3 objects