DNA Weaver (documentation here) is a Python library to find optimal strategies for assembling large DNA constructs. Given an arbitrary sequence, DNA Weaver will select the most adapted commercial DNA providers, cloning methods and parts repositories (depending on your preferences), and will design all necessary assembly fragments and assembly steps. Try it online via the DNA Weaver web app.
DNA Weaver was written with versatility and extensibility in mind: each DNA source and assembly method can be customized, and assembly plans can be optimized with respect to total price, overall duration of the assembly, or assembly success probabilities.
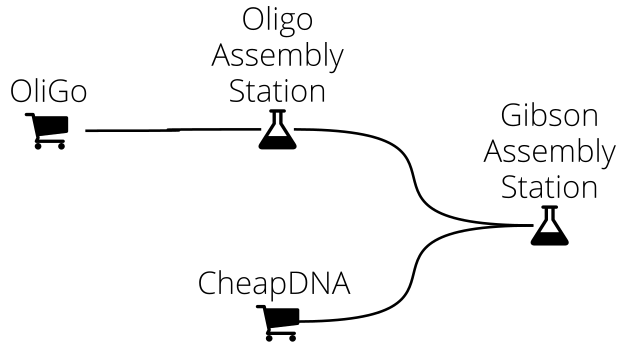
In DNA Weaver you first define a supply network connecting various DNA sources (commercial providers, part repositories, genomic DNA, and cloning stations) to represent how DNA can be obtained in your lab or biofoundry. For instance, assume that you routinely assemble ~10kb sequences using Gibson assembly, from fragments obtained either commercially or from the assembly of oligonucleotides. Your supply network then looks as follows:
When you submit a sequence to the main station (here, the Gibson Assembly station), DNA Weaver will use smart sequence decomposition techniques and competitive bidding between the different DNA sources in order to find the best possible assembly plan.
A detailed description and walkthrough of DNA Weaver can be found in this PDF document: DNA Weaver presentation.
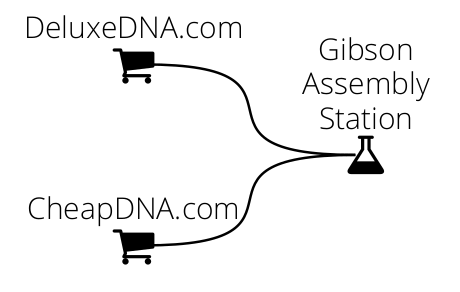
In the following example we ask DNA Weaver for a plan to assemble a 10kb sequence via Gibson assembly of fragments with 40bp homologies. The fragments can be ordered from two companies: CheapDNA produces fragments for 10c/bp, at the condition that they do not contain any BsaI site and are smaller than 3kb, and DeluxeDNA produces produces any fragment under 4kb for 20c/bp.
We will first CheapDNA and DeluxeDNA separately, then link them to a Gibson Assembly station:
import dnaweaver as dw
cheap_dna_offer = dw.CommercialDnaOffer(
name="CheapDNA",
sequence_constraints=[
dw.NoPatternConstraint(enzyme="BsaI"),
dw.SequenceLengthConstraint(max_length=4000)
],
pricing=dw.PerBasepairPricing(0.10),
lead_time=40
)
deluxe_dna_offer = dw.CommercialDnaOffer(
name="DeluxeDNA",
sequence_constraints=[dw.SequenceLengthConstraint(max_length=3000)],
pricing=dw.PerBasepairPricing(0.20),
lead_time=20
)
assembly_station = dw.DnaAssemblyStation(
name="Gibson Assembly Station",
assembly_method=dw.GibsonAssemblyMethod(
overhang_selector=dw.TmSegmentSelector(min_tm=55, max_tm=70),
min_segment_length=500,
max_segment_length=4000,
duration=5
),
supplier=[cheap_dna_offer, deluxe_dna_offer],
coarse_grain=20
)
sequence = dw.random_dna_sequence(10000, seed=123)
quote = assembly_station.get_quote(sequence, with_assembly_plan=True)
print (quote.assembly_step_summary())This code prints out an assembly summary showing the source of the different sequence segments (start, end):
Ordering plan:
0-1719: From CheapDNA - price 172.80 - lead_time 40.0
1719-4429: From CheapDNA - price 273.00 - lead_time 40.0
4429-5318: From DeluxeDNA - price 182.00 - lead_time 20.0
5318-7359: From CheapDNA - price 206.00 - lead_time 40.0
7359-10000: From CheapDNA - price 265.00 - lead_time 40.0
Price: 1098.80, total lead_time: 45.0Notice how DNA Weaver uses preferentially CheapDNA, with the exception of a 1kb fragment in the middle of the sequence, which had to be ordered from DeluxeDNA due to the presence of a BsaI site.
By defining more DNA sources and connecting them together it is possible to model complex assembly problems.
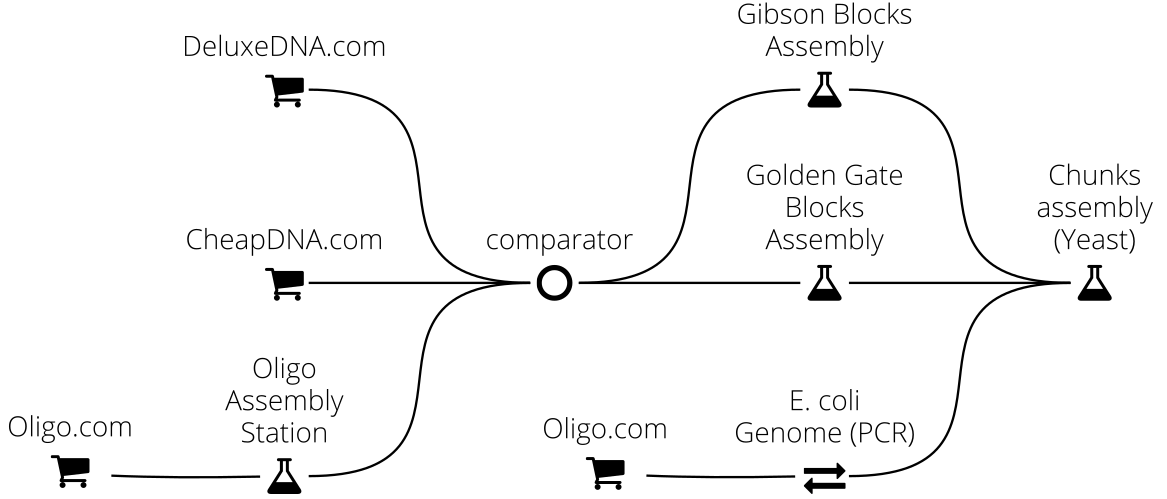
For instance in this example we implement a complex DNA assembly chain, where the final DNA sequence (typically 50kb) is obtained from Yeast recombination of DNA chunks originating either from the E. coli chromosome (via PCR extraction) or from the assembly of smaller fragments via Golden Assembly or Gibson assembly (whichever method is best adapted). These assembly fragments are obtained either from commercial providers (CheapDNA and DeluxeDNA) or assembled from oligos:
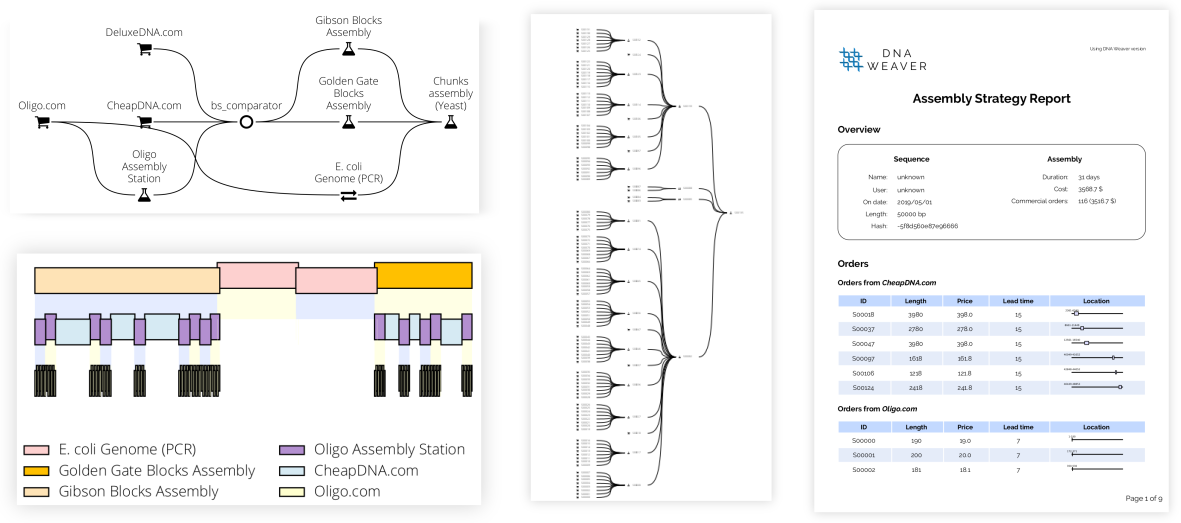
Just a few lines of code can produce a comprehensive report (see a sample here) featuring plots of the final assembly plan, comprehensive PDF reports listing all operations needed, and genbank/fasta files of the sequences to order:
quote = assembly_station.get_quote(sequence, with_assembly_plan=True)
assembly_plan_report = quote.to_assembly_plan_report()
assembly_plan_report.write_full_report("report.zip")Result:
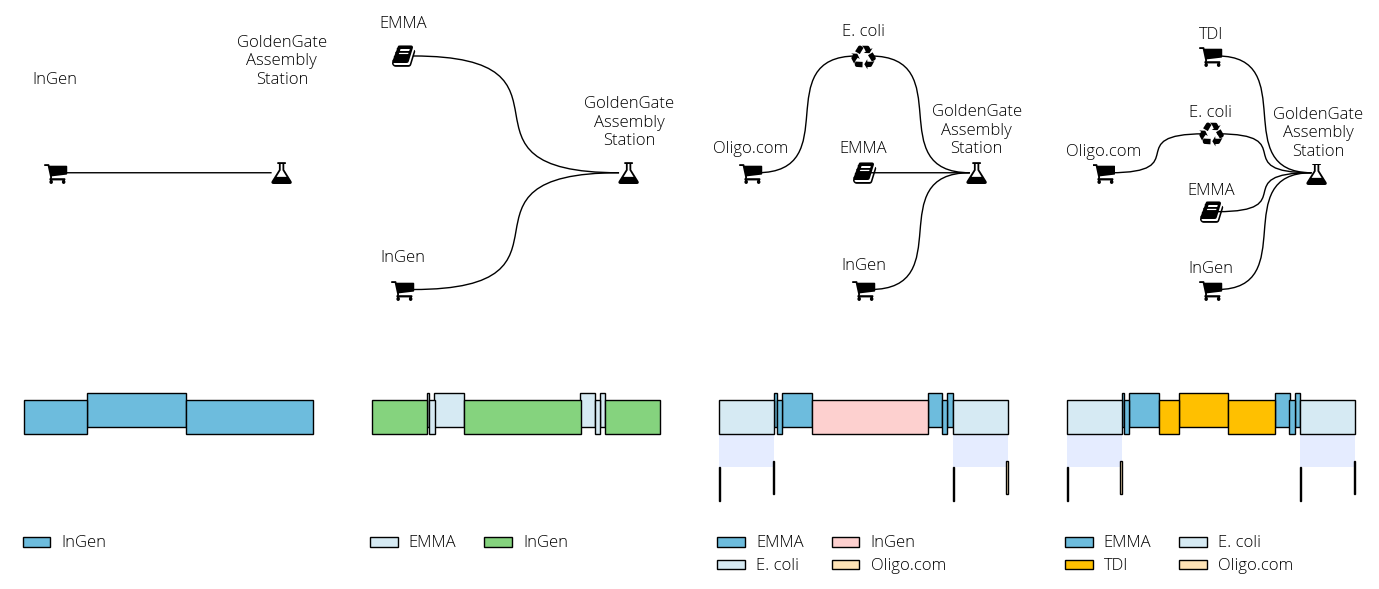
In this other example we build a sequence comprising a resistance cassette (promoter, resistance, terminator) flanked by two homology arms. The sequence incorporates parts from the EMMA library. The script progressively adds new DNA sources (commercial DNA, the EMMA library, chromosomal DNA) so we can observe the changes in the proposed solution:
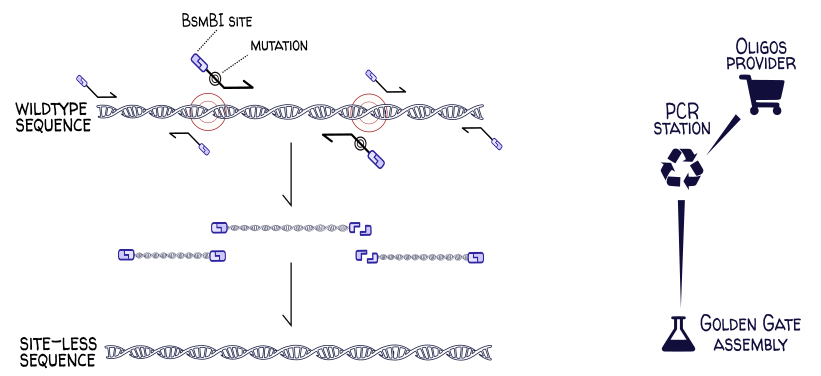
A common cloning operation is the domestication of a genetic part for a given assembly standard. Many Golden Gate assembly standards forbid BsaI and BsmBI restriction sites in part sequences. If one wanted to use the wildtype E. coli gene yeeJ, one would need to first remove the BsaI and BsmBI sites at positions 453, 2284, 3979, 5455 and 5990 in the gene sequence. This can be done via site-directed mutagenesis, where regions of the chromosome are PCR-amplified at precise locations using carefully-designed primers. These primers have overhangs introducing the desired (codon-synonymous) mutations and (in this example) carry BsaI sites so that the PCR products can be digested and assembled into the site-less final sequence.
This process can be easily modeled in DNA Weaver by connecting a PCR station (and its oligo provider) to an assembly station:
import dnaweaver as dw
oligos_company = dw.CommercialDnaOffer(
"OligoCompany",
sequence_constraints=[dw.SequenceLengthConstraint(max_length=200)],
pricing=dw.PerBasepairPricing(0.1)
)
pcr_station = dw.PcrExtractionStation(
name="PCR station",
max_overhang_length=50,
primers_supplier=oligos_company,
blast_database='./ecoli_genome/ecoli',
extra_cost=5
)
assembly_station = dw.DnaAssemblyStation(
name="Golden Gate assembly",
assembly_method = dw.GoldenGateAssemblyMethod(enzyme='BsaI'),
supplier=pcr_station,
coarse_grain=100,
fine_grain=0,
logger='bar'
)
# LOAD THE (SITE-FREE) DESIRED SEQUENCE
desired_sequence = str(dw.load_record("./desired_sequence.gb").seq)
# THIS LINE WILL PRE-BLAST THE SEQUENCE TO ACCELERATE COMPUTATIONS.
assembly_station.prepare_network_on_sequence(desired_sequence)
# FIND AN ASSEMBLY PLAN AND PRINT IT.
quote = assembly_station.get_quote(desired_sequence)
print (quote.assembly_step_summary())Result:
Ordering plan: 0-451: From PCR station - price 11.70 - lead_time 0.0 - From gnl|BL_ORD_ID|0_h000_00 451-2283: From PCR station - price 12.60 - lead_time 0.0 - From gnl|BL_ORD_ID|0_h000_01 2283-3987: From PCR station - price 12.00 - lead_time 0.0 - From gnl|BL_ORD_ID|0_h000_02 3987-5451: From PCR station - price 11.80 - lead_time 0.0 - From gnl|BL_ORD_ID|0_h000_03 5451-5985: From PCR station - price 11.80 - lead_time 0.0 - From gnl|BL_ORD_ID|0_h000_04 5985-7077: From PCR station - price 11.90 - lead_time 0.0 - From gnl|BL_ORD_ID|0_h000_05 Price:71.80, total lead_time:0.0
The full assembly report (which you can generate in this example) has the list of all primers to order (including overhangs with sequence mutations and BsaI sites).
You can install DnaWeaver through PIP:
pip install dnaweaver
Alternatively, you can unzip the sources in a folder and type:
python setup.py install
Also install the ncbi-blast+ package to be able to use PCR stations. On Ubuntu:
sudo apt-get install ncbi-blast+
You may also need the following non-Python dependencies for report generation. On Ubuntu:
sudo apt-get install build-essential python3-dev python3-pip \
python3-cffi libcairo2 libpango-1.0-0 libpangocairo-1.0-0 \
libgdk-pixbuf2.0-0 libffi-dev shared-mime-info
DNA Weaver is an open-source software originally written at the Edinburgh Genome Foundry by Zulko and released on Github under the MIT licence (Copyright 2017 Edinburgh Genome Foundry).
Everyone is welcome to contribute!

DNA Weaver is part of the EGF Codons synthetic biology software suite for DNA design, manufacturing and validation.