⚠️ This repo is no longer maintained! Please check out our brand new SuperPoint Transformer, which does everything better! ⚠️
This is the official PyTorch implementation of the papers:
Large-scale Point Cloud Semantic Segmentation with Superpoint Graphs
by Loic Landrieu and Martin Simonovski (CVPR2018),
and
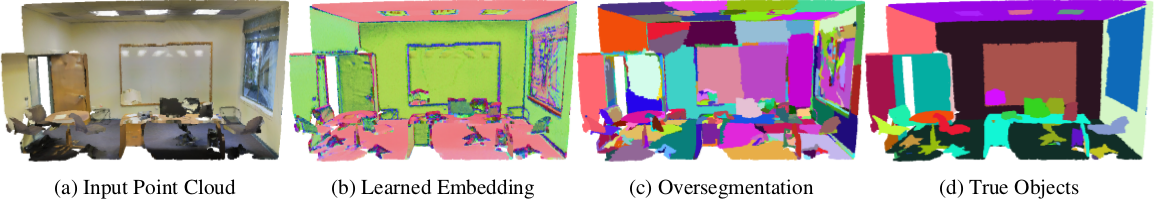
Point Cloud Oversegmentation with Graph-Structured Deep Metric Learning.
by Loic Landrieu and Mohamed Boussaha (CVPR2019),
./partition/*- Partition code (geometric partitioning and superpoint graph construction using handcrafted features)./supervized_partition/*- Supervized partition code (partitioning with learned features)./learning/*- Learning code (superpoint embedding and contextual segmentation).
To switch to the stable branch with only SPG, switch to release.
Our partition method is inherently stochastic. Hence, even if we provide the trained weights, it is possible that the results that you obtain differ slightly from the ones presented in the paper.
0. Download current version of the repository. We recommend using the --recurse-submodules option to make sure the cut pursuit module used in /partition is downloaded in the process. Wether you did not used the following command, please, refer to point 4:
git clone --recurse-submodules https://github.com/loicland/superpoint_graph
1. Install PyTorch and torchnet.
pip install git+https://github.com/pytorch/tnt.git@master
2. Install additional Python packages:
pip install future igraph tqdm transforms3d pynvrtc fastrlock cupy h5py sklearn plyfile scipy pandas
3. Install Boost (1.63.0 or newer) and Eigen3, in Conda:
conda install -c anaconda boost; conda install -c omnia eigen3; conda install eigen; conda install -c r libiconv
4. Make sure that cut pursuit was downloaded. Otherwise, clone this repository or add it as a submodule in /partition:
cd partition
git submodule init
git submodule update --remote cut-pursuit
5. Compile the libply_c and libcp libraries:
CONDAENV=YOUR_CONDA_ENVIRONMENT_LOCATION
cd partition/ply_c
cmake . -DPYTHON_LIBRARY=$CONDAENV/lib/libpython3.6m.so -DPYTHON_INCLUDE_DIR=$CONDAENV/include/python3.6m -DBOOST_INCLUDEDIR=$CONDAENV/include -DEIGEN3_INCLUDE_DIR=$CONDAENV/include/eigen3
make
cd ..
cd cut-pursuit
mkdir build
cd build
cmake .. -DPYTHON_LIBRARY=$CONDAENV/lib/libpython3.6m.so -DPYTHON_INCLUDE_DIR=$CONDAENV/include/python3.6m -DBOOST_INCLUDEDIR=$CONDAENV/include -DEIGEN3_INCLUDE_DIR=$CONDAENV/include/eigen3
make
6. (optional) Install Pytorch Geometric
The code was tested on Ubuntu 14 and 16 with Python 3.5 to 3.8 and PyTorch 0.2 to 1.3.
Common sources of errors and how to fix them:
- $CONDAENV is not well defined : define it or replace $CONDAENV by the absolute path of your conda environment (find it with
locate anaconda) - anaconda uses a different version of python than 3.6m : adapt it in the command. Find which version of python conda is using with
locate anaconda3/lib/libpython - you are using boost 1.62 or older: update it
- cut pursuit did not download: manually clone it in the
partitionfolder or add it as a submodule as proposed in the requirements, point 4. - error in make:
'numpy/ndarrayobject.h' file not found: set symbolic link to python site-package withsudo ln -s $CONDAENV/lib/python3.7/site-packages/numpy/core/include/numpy $CONDAENV/include/numpy
To run our code or retrain from scratch on different datasets, see the corresponding readme files. Currently supported dataset are as follow:
| Dataset | handcrafted partition | learned partition |
|---|---|---|
| S3DIS | yes | yes |
| Semantic3D | yes | to come soon |
| vKITTI3D | no | yes |
| ScanNet | to come soon | to come soon |
To use pytorch-geometric graph convolutions instead of our own, use the option --use_pyg 1 in ./learning/main.py. Their code is more stable and just as fast. Otherwise, use --use_pyg 0
To evaluate quantitatively a trained model, use (for S3DIS and vKITTI3D only):
python learning/evaluate.py --dataset s3dis --odir results/s3dis/best --cvfold 123456
To visualize the results and all intermediary steps, use the visualize function in partition (for S3DIS, vKITTI3D,a nd Semantic3D). For example:
python partition/visualize.py --dataset s3dis --ROOT_PATH $S3DIR_DIR --res_file results/s3dis/pretrained/cv1/predictions_test --file_path Area_1/conferenceRoom_1 --output_type igfpres
output_type defined as such:
'i'= input rgb point cloud'g'= ground truth (if available), with the predefined class to color mapping'f'= geometric feature with color code: red = linearity, green = planarity, blue = verticality'p'= partition, with a random color for each superpoint'r'= result cloud, with the predefined class to color mapping'e'= error cloud, with green/red hue for correct/faulty prediction's'= superedge structure of the superpoint (toggle wireframe on meshlab to view it)
Add option --upsample 1 if you want the prediction file to be on the original, unpruned data (long).
You can apply SPG on your own data set with minimal changes:
- adapt references to
custom_datasetin/partition/partition.py - you will need to create the function
read_custom_formatin/partition/provider.pywhich outputs xyz and rgb values, as well as semantic labels if available (already implemented for ply and las files) - adapt the template function
/learning/custom_dataset.pyto your achitecture and design choices - adapt references to
custom_datasetin/learning/main.py - add your data set colormap to
get_color_from_labelin/partition/provider.py - adapt line 212 of
learning/spg.pyto reflect the missing or extra point features - change
--model_configtogru_10,f_KwithKas the number of classes in your dataset, orgru_10_0,f_Kto use matrix edge filters instead of vectors (only use matrices when your data set is quite large, and with many different point clouds, like S3DIS).
If your data does not have RGB values you can easily use SPG. You will need to follow the instructions in partition/partition.ply regarding the pruning.
You will need to adapt the /learning/custom_dataset.py file so that it does not refer ro RGB values.
You should absolutely not use a model pretrained on values with RGB. instead, retrain a model from scratch using the --pc_attribs xyzelpsv option to remove RGB from the shape embedding input.
If you use the semantic segmentation module (code in /learning), please cite:
Large-scale Point Cloud Semantic Segmentation with Superpoint Graphs, Loic Landrieu and Martin Simonovski, CVPR, 2018.
If you use the learned partition module (code in /supervized_partition), please cite:
Point Cloud Oversegmentation with Graph-Structured Deep Metric Learning, Loic Landrieu and Mohamed Boussaha CVPR, 2019.
To refer to the handcrafted partition (code in /partition) step specifically, refer to:
Weakly Supervised Segmentation-Aided Classification of Urban Scenes from 3D LiDAR Point Clouds, Stéphane Guinard and Loic Landrieu. ISPRS Workshop, 2017.
To refer to the L0-cut pursuit algorithm (code in github.com/loicland/cut-pursuit) specifically, refer to:
Cut Pursuit: Fast Algorithms to Learn Piecewise Constant Functions on General Weighted Graphs, Loic Landrieu and Guillaume Obozinski, SIAM Journal on Imaging Sciences, 2017
To refer to pytorch geometric implementation, see their bibtex in their repo.